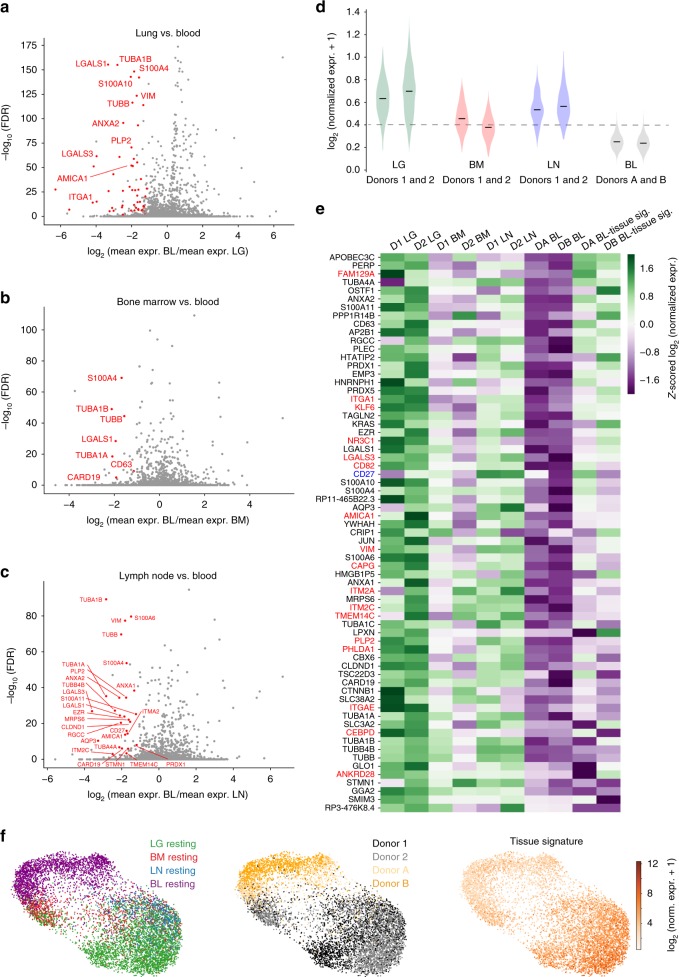
Fig. 3.
Identification of a tissue gene signature for resting memory T cells. a Volcano plot showing the average log-fold-change and average Benjamini-Hochberg-corrected p-values (FDR) for pairwise differential expression between CCL5+ T cells from each resting LG sample and each resting blood sample. Genes with negative log-fold-change are more highly expressed among CCL5+ cells in LG, with several differentially expressed genes (multiple test-corrected Wilcoxon p < 0.05, fold change >2) highlighted in red. b Same as (a) for comparison of resting CCL5+ T cells in BM and blood. c Same as (a) for comparison of resting CCL5+ T cells in LN in blood. d Violin plot showing the distributions of the average expression of all genes with two-fold higher expression (on average) in any tissue compared to blood and average FDR < 0.05 (described above) in any tissue for the resting CCL5+ T cells in each tissue and blood sample. The dashed line marks one standard deviation below the mean for average expression of this signature for all tissues (note a small number of blood cells fall above this line). e Heatmap shows z-scored average expression for all genes in the tissue signature from (d) among the resting CCL5+ T cells from each tissue and blood sample plus that of the rare blood subpopulation from (d), which expresses high levels of a subset of tissue signature genes. Previously identified TRM-associated genes from bulk RNA-seq studies are highlighted in red (enriched in CD69+ vs. CD69− tissue memory T cells)13, and CD27 highlighted in blue was previously found to be upregulated on human TRM compared to TEM cells36. f UMAP embedding of resting CCL5+ T cells (TEM cells) from all four donors generated using the tissue-associated T cell signature colored by tissue site (left), donor (center), and average expression of the signature (right). Source data listing genes and expression values for (a–e) are provided in the Source Data file