Fig. 4.
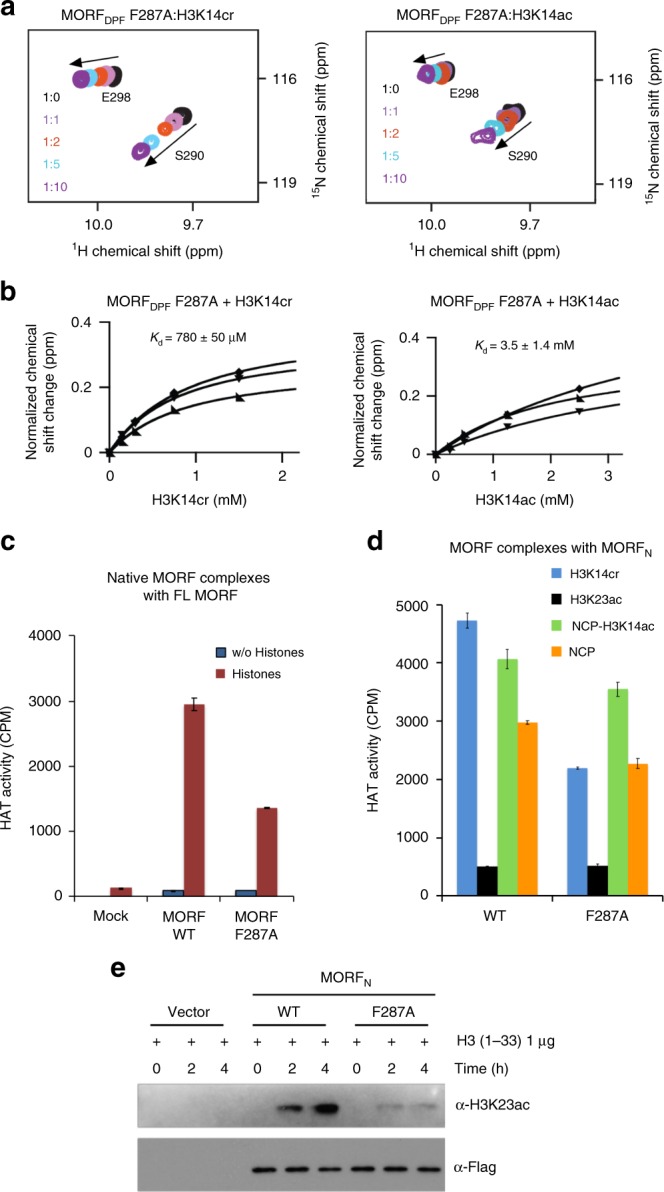
H3K23 acetylation depends on functional MORFDPF. a Superimposed 1H,15N HSQC spectra of MORFDPF F287A mutant collected as indicated peptides were added stepwise. Spectra are color coded according to the protein:peptide molar ratio (inset). b Representative binding curves used to determine Kds for the interactions of MORFDPF F287A with indicated peptides by NMR. Error was calculated as SD. c HAT activity of native MORF wild-type and F287A mutant complexes from K562 cells on native free core histones. Liquid HAT assays, in which reactions were spotted on P81 filters and counted by scintillation as counts per minute (CPM). d HAT activities of wild-type and mutated MORFN complexes purified from 293T cells using indicated H3 peptides (residues 1–29) or recombinant unmodified nucleosomes (NCP) or nucleosomes containing H3K14ac (H3K14ac-NCP) as substrates. Error bars in c and d indicate the range from duplicate samples, n = 2 independent experiments. e Western blot analysis of HAT assays with purified overexpressed wild-type and mutant MORFN using H3 peptide (residues 1–33) as substrate
