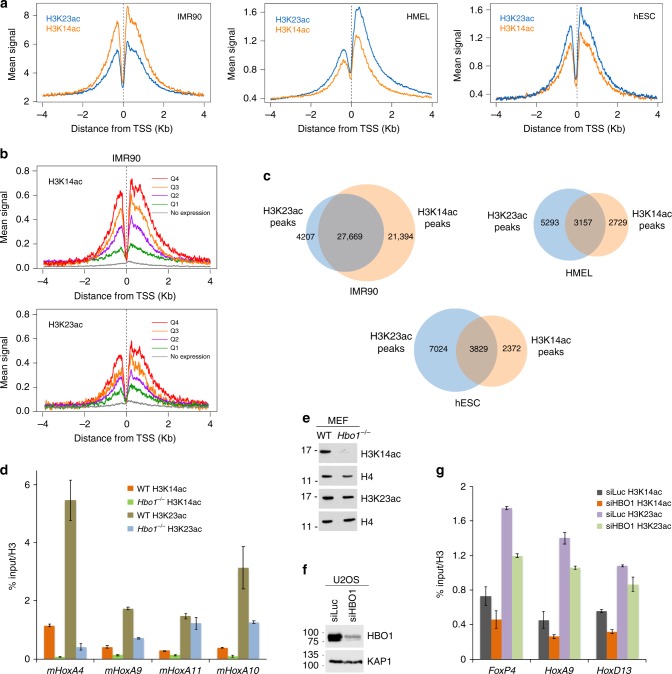
Fig. 7.
H3K23ac and H3K14ac colocalization and effect of H3K14ac on H3K23 acetylation at specific genes. a Metagene analysis of H3K14ac and H3K23ac signals around the transcription start sites of genes observed in ChIP-seq datasets derived from ENCODE in IMR90, hESC and HMEL cells. b RNA-seq analysis of H3K14ac and H3K23ac in IMR90 cell line derived from ENCODE. Genes were subdivided in quartiles corresponding to their expression levels. c Venn diagrams show overlap of the H3K14ac and H3K23ac peaks observed in IMR90, hESC and HMEL cells. d ChIP analysis of H3K23ac and H3K14ac in WT or Hbo1−/− MEFs. Acetylation levels were corrected for nucleosomes occupancy (total H3 signal). e Western blot analysis of histones purified from WT and Hbo1−/− MEFs with indicated antibodies. f Western blot analysis of HBO1 and KAP1 in U2OS cells treated with siLuciferase or siHBO1. g ChIP analysis of H3K23ac and H3K14ac in U2OS cells treated with siLuciferase or siHBO1. Acetylation levels were corrected for nucleosomes occupancy (total H3 signal). All ChIP values are based on two independent experiments and error bars in d and g indicate the range from duplicate samples, n = 2 independent experiments. Source data are provided as a Source Data file