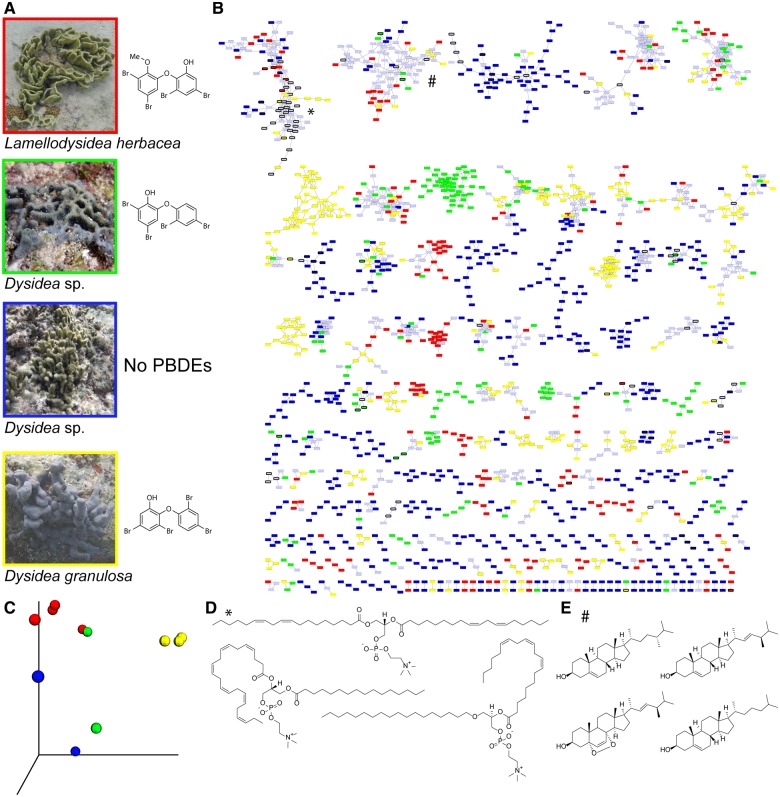
Fig. 1.
Metabolomics for Dysideidae sponges. (A) Four representative Dysideidae sponges collected by the authors in Guam (years 2014–2015) with the dominant PBDEs present in each sponge. (B) A molecular network of metabolites detected in the extracts of Dysideidae sponges. Nodes unique to a Dysideidae species are color-coded as in panel A. Nodes shared between multiple species are colored gray. A molecular network was created using the online workflow at GNPS. The data were filtered by removing all MS/MS peaks within ±17 Da of the precursor m/z. The data were then clustered with MS-Cluster with a parent mass tolerance of 0.1 Da and a MS/MS fragment ion tolerance of 0.1 Da to create consensus spectra. Further, consensus spectra that contained less than 1 spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 4 matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 4 matched peaks. Multiple biological replicates were used for each sponge species. (C) Principal component analyses of the sponge metabolomes. Biological replicates of Lamellodysidea herbacea (in red) and of Dysidea granulosa (yellow) cluster together and can be neatly differentiated. A greater divergence is observed for the other two Dysidea sp. sponges which can be resolved by increasing the number of replicates in this analysis. Dereplicated (D) phospholipids (denoted by * in panel B) and (E) sterols (denoted by # in panel B) in the Dysideidae molecular network. Chemical identities of a large majority of Dysideidae metabolites remain unknown.