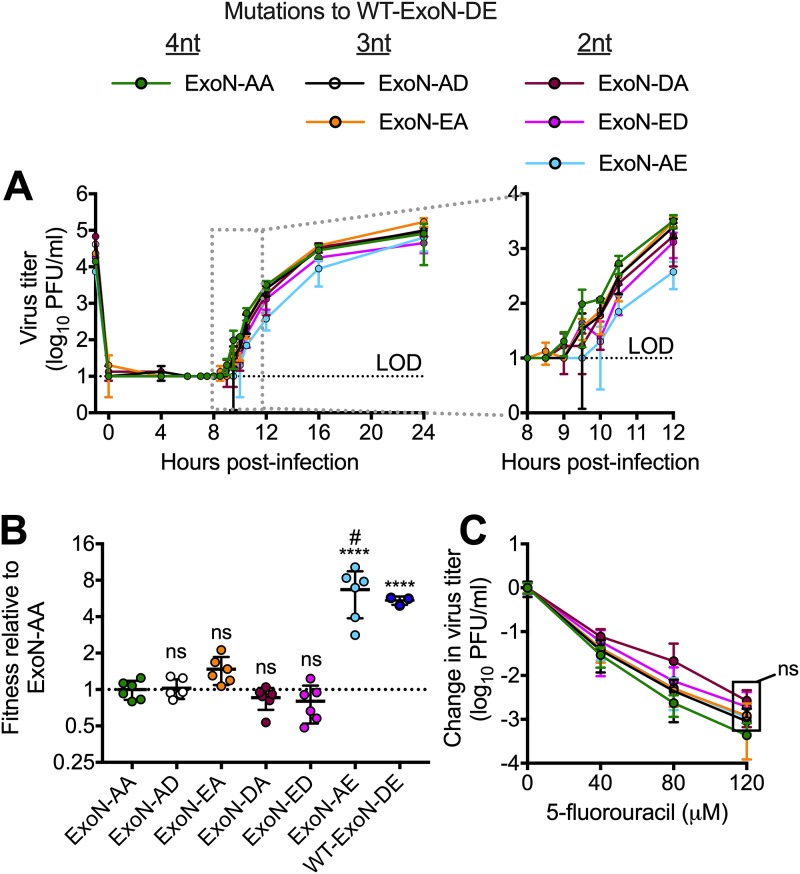
FIG 2.
Intermediate revertants of ExoN-AA motif I do not have selective advantages. (A) Replication kinetics at an MOI of 0.01 PFU/cell plotted as means ± standard deviations (n = 3). (B) Competitive fitness of each variant relative to that of ExoN-AA. Viruses were competed with a tagged ExoN-AA-reference strain, and relative fitness was normalized to the mean of the ExoN-AA value. (C) 5-Fluorouracil sensitivity at an MOI of 0.01 PFU/cell. Statistical significance of each variant relative to the value of ExoN-AA was determined by one-way analysis of variance with multiple comparisons (D) and by two-way analysis of variance with Dunnett’s multiple-comparison test (C). ****, P < 0.0001; ns, not significant. LOD, limit of detection. Data in panels B and C represent means ± standard deviations (n = 6). Boxed values have the same significance. #, all lineages of ExoN-AE reverted to WT-ExoN-DE during the experiment.