Table 2.
ϕ‐Value analysis of coupled folding‐binding process
| IDP | Target | ϕ‐Value distribution | Reference | IDP | Target | ϕ‐Value distribution | Reference |
|---|---|---|---|---|---|---|---|
| HIF‐1α | TAZ1 |
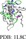
|
137 | c‐Myb | KIX |

|
88, 138, 139 |
| STAT2 | TAZ1 |

|
96 | pKID | KIX |

|
140 |
| PUMA | A1 |

|
141 | MLL | KIX |

|
142 |
| PUMA | MCL‐1 |

|
130, 141 | E6 peptide | PDZ2 |

|
143 |
| BID | A1 |

|
141 | S peptide | S protein |

|
144 |
| BID | MCL‐1 |

|
141 | NTAIL | X domain |

|
145 |
| α‐Spectrin | β‐Spectrin |

|
146 | ACTR | NCBD |

|
86, 147, 148 |
| C‐terminal tail of nNOS PDZ | Syntrophin PDZ |

|
149 |
Note: The IDPs are colored in light green and the targets are colored in gray. Residues with low (ϕ ≤ 0.25), medium (0.25 < ϕ < 0.6), and high (ϕ ≥ 0.6) ϕ‐values are highlighted in blue, magenta, and red, respectively.
