Abstract
Objective
Electronic health records linked with biorepositories are a powerful platform for translational studies. A major bottleneck exists in the ability to phenotype patients accurately and efficiently. The objective of this study was to develop an automated high-throughput phenotyping method integrating International Classification of Diseases (ICD) codes and narrative data extracted using natural language processing (NLP).
Materials and Methods
We developed a mapping method for automatically identifying relevant ICD and NLP concepts for a specific phenotype leveraging the Unified Medical Language System. Along with health care utilization, aggregated ICD and NLP counts were jointly analyzed by fitting an ensemble of latent mixture models. The multimodal automated phenotyping (MAP) algorithm yields a predicted probability of phenotype for each patient and a threshold for classifying participants with phenotype yes/no. The algorithm was validated using labeled data for 16 phenotypes from a biorepository and further tested in an independent cohort phenome-wide association studies (PheWAS) for 2 single nucleotide polymorphisms with known associations.
Results
The MAP algorithm achieved higher or similar AUC and F-scores compared to the ICD code across all 16 phenotypes. The features assembled via the automated approach had comparable accuracy to those assembled via manual curation (AUCMAP 0.943, AUCmanual 0.941). The PheWAS results suggest that the MAP approach detected previously validated associations with higher power when compared to the standard PheWAS method based on ICD codes.
Conclusion
The MAP approach increased the accuracy of phenotype definition while maintaining scalability, thereby facilitating use in studies requiring large-scale phenotyping, such as PheWAS.
Keywords: High-throughput, phenotyping, PheWAS
INTRODUCTION
High-throughput technologies have provided powerful tools for dissecting the genomic and biologic architecture of complex human traits. To continue on the trajectory of their success, more work is needed to translate ‘omics’ findings to improvement in patient care. A major challenge in realizing such translation is linking the high-throughput biologic data with detailed high-quality phenotypic data. To improve their translational potential, ‘omics’ studies are increasingly conducted using cohorts with linked electronic health records (EHR) data and biobanks. Such studies are typically designed to investigate hundreds to thousands of phenotypes simultaneously, thereby identifying a large unmet need for scalable, standardized, efficient, and portable approaches for phenotyping.
The current standard for studying hundreds to thousands of phenotypes over millions of patients is the use of diagnoses codes such as the International Classification of Diseases (ICD) codes. The phenome-wide association studies (PheWAS) approach1 was developed specifically for use in EHRs linked with biobanks to screen for associations between a genetic marker with roughly 1800 phenotypes. The phenotypes in the PheWAS are generated by grouping ICD-9 codes. However, a known pitfall of ICD codes are variations in accuracy, leading to loss of power for association studies.2–5 Rule-based manual curation and machine learning-based supervised learning approaches have been proposed as more accurate alternatives for phenotyping and have been used to create EHR-based patient cohorts for discovery research.6–18 These approaches enhance the accuracy of phenotyping by combining information from multiple EHR features, including codified features such as ICD codes and narrative features identified via natural language processing (NLP). While collaborative platforms such as the PheKB19 share existing phenotyping algorithms, deploying these algorithms across institutions can be labor intensive and limiting the feasibility of using these algorithms for phenotype screens.
To improve the efficiency for developing phenotyping algorithms, various approaches have been developed to reduce the level of human input required. Examples of these approaches include active sampling, feature refinement, and feature selection.20–23 Several automated annotation methods have also been proposed24,25; however, these methods either still require expert input to curate silver standard labels or have variable accuracy. Recently, Yu et al developed PheNorm,26 a fully automated phenotyping algorithm based on normal mixture modeling using ICD codes only. While PheNorm presented a highly scalable approach by ranking participants with respect to their likelihood of having a phenotype, it could not provide the final classification of whether a participant had a specific phenotype necessary for clinical studies.
In this article, we propose an unsupervised multimodal automated phenotyping (MAP) method with application in PheWAS. We hypothesize that MAP, which incorporates NLP, will efficiently and accurately classify millions of participants across the roughly 1800 phenotypes in the PheWAS compared to existing approaches.
MATERIALS AND METHODS
The MAP procedure includes 2 key steps: (1) assembling the ICD codes and NLP features corresponding to the target phenotype, and (2) annotation via unsupervised ensemble latent mixture modeling.
Identifying the main ICD and NLP features for phenotypes
For a target phenotype, we first identify the main ICD code associated with the phenotype. The main ICD code is either defined by the investigator or by selecting a PheWAS code (phecode) from the catalog (https://phewascatalog.org/phecodes) to use the associated ICD mapping.1,27 Briefly, the phecode is a published mapping of grouping ICD codes into clinically relevant groups. For example, salmonella pneumonia (PNA) (ICD-9 003.22), PNA due to streptococcus (ICD-9 482.3), etc., are grouped into1 phecode “480.1” for “bacterial pneumonia.” Multiple or higher level phecodes can also be used together to represent a broader category (eg, diabetes mellitus [phecode 250] which includes type 1 diabetes [phecode 250.1] and type 2 diabetes [phecode 250.2]). The PheWAS catalog provides a hierarchical roll-up of information to aggregate broader phenotypes. For a given list of ICD-9 codes selected via phecode mapping or domain expert to represent the phenotype, all codes are aggregated to represent the main ICD feature. Next, the total count of the main ICD feature is extracted from the data set, denoted by . All ICD-10 codes were first mapped to ICD-9 codes based on the General Equivalence Mapping (GEMS) available at the Centers for Medicare & Medicaid Services.28 Each ICD-9 code is only counted once per day to reduce the impact of dual coding.
The main NLP feature used in MAP for the phenotype is determined through identifying the medical concepts by mapping relevant clinical terms to the concept unique identifiers (CUIs) listed in the Unified Medical Language System (UMLS). While the main medical concept can be selected by a domain expert, we find it more robust to identify the CUIs through a mapping process utilizing the PheWAS catalog map, in which a phenotype text string is used to describe each phecode. For each phecode, we identify its corresponding ICD codes along with the phenotype string. We identify a list of CUIs for the phecode through 3 mapping steps: (1) mapping all relevant ICD-9 codes directly to CUIs, which can be performed directly in UMLS using the (CODE field in the MRCONSO table); (2) mapping ICD-9 strings to CUIs (using the STR field in the MRCONSO table) with exact string-matching; and (3) mapping the phenotype string to CUIs (again using the MRCONSO STR field) with exact string-matching (Supplementary Figure 1). Although steps (1) and (2) typically yield identical lists, (2) occasionally results in a larger list since an ICD-9 string may be mapped to multiple concepts. Combining all CUIs mapped in the 3 steps gives a list of CUIs to represent each phecode.
The narrative text notes for all participants are then processed. Using the CUI list assembled for each phecode, the total number of positive mentions of any clinical terms belonging to the CUIs list, denoted by , is used as the main NLP feature for the phecode. If a phenotype is defined by several phecodes, we take the sum of the corresponding NLP counts as the feature for the phenotype. The Narrative Information Linear Extraction (NILE)29 was used to extract concepts in the experiments of this study. Based on previous phenotyping studies, the NLP features extracted were similar regardless of NLP platform used.10–18
As demonstrated in the Yu et al26 PheNorm study, the level of health care utilization contributed noise to algorithm features and dampened their ability to predict the phenotype status. We thus include the total number of narrative notes for each patient, denoted by , as a proxy for the health care utilization in the MAP algorithm. We also consider modeling the logarithm transformed features, , and . Additionally, similar to Yu et al,26 we combine ICD and NLP counts to create additional features and .
Unsupervised MAP prediction
Using patient level data on the , , ,,, , and, we separately fitted mixture models to each individual feature along with to obtain the probability that a patient has the phenotype. Specifically, for , we fitted a Poisson mixture model with , where is the unobserved phenotype status. The probability mass function of is, therefore,
where the parameters , , , and are estimated with the expectation-maximization (EM) algorithm. The parameter is the prevalence of the phenotype, is essentially the health care utilization adjusted mean of among those with for , and is used to mitigate the noise brought into in predicting the phenotype status, where is generally negative. To increase the robustness of the procedure, we also fit normal mixture models to the log-transformed count data with whose probability density function is
where , , , , , and are similarly estimated with the EM algorithm. The parameters and respectively denote the health care utilization adjusted mean and variance of among those with phenotype . We fit the above Poisson and log-normal mixture models to the ICD and NLP features, as either or both models may provide a good approximation to the observed data, and it is unclear which model provides a better approximation for a given data set—especially in the absence of gold standard labels on .
To assign a predicted probability to each participant, we calculate the posterior probabilities of having the phenotype given the feature information from each fitted mixture model via the Bayes’ rule. Under each model, the prevalence of the phenotype can be estimated by the average value of the posterior probabilities across all patients. The final MAP algorithm assigns the predicted probability of having the phenotype as the average of the 6 predicted probabilities. We also estimate the prevalence of the phenotype by averaging the prevalence estimates from the 6 latent mixture model fittings. The prevalence estimate is then used as a threshold to assign a binary classification of whether a participant has the phenotype. Participants with predicted probabilities above are assigned as having the phenotype with , and those below are assigned as not having the phenotype with (see Supplementary Method for detailed MAP algorithm description).
Validation of MAP using real-world biorepository data
Study populations
Partners Biobank contains linked EHR and genetic data anchored by 2 large tertiary care hospitals: Brigham and Women’s Hospital and Massachusetts General Hospital in Boston. A total of 17 805 patients had codified data (NLP data as well as genetic data).30,31 Additionally, gold standard labels were curated on an average of 545 patients for 16 phenotypes: asthma, bipolar disorder (BD), schizophrenia (SCZ), breast cancer (BrCa), chronic obstructive pulmonary disease (COPD), congestive heart failure (CHF), coronary artery disease (CAD), hypertension (HTN), depression (DPRSSN), epilepsy, multiple sclerosis (MS), rheumatoid arthritis (RA), type I diabetes mellitus, type II diabetes mellitus, Crohn’s disease (CD), and ulcerative colitis (UC).
The Veteran’s Affairs Million Veteran’s Project (VA MVP) is a longitudinal cohort study, recruiting at approximately 50 VA facilities in the United States.32 MVP data was used to validate the MAP approach, and to demonstrate the application of MAP to a PheWAS. A total of 330 374 patients had both EHR and genetic data available for the PheWAS analysis.
Overview of analyses
The performance of the MAP algorithm was validated in 3 separate experiments: (1) phenotyping 16 conditions using MAP compared against the gold-standard labels obtained from chart review in the Partners Biobank; (2) an association study between a low-density lipoprotein cholesterol (LDL-C) genetic risk score (GRS) and hyperlipidemia phenotypes; and (3) a PheWAS in MVP of the interleukin-6R (IL6R) genetic variant (Asp358Ala, rs2228145).33
Testing the performance of MAP against existing approaches for phenotyping across 16 conditions with gold standard labels
In the Partners Biobank, we compared the accuracy of (1) the main ICD feature, (2) the main NLP feature, (3) the PheNorm algorithm, and (4) the MAP algorithm. Similar to previous phenotyping approaches,21,22,26,31 we first applied a filter to create a data set of participants who may have the phenotype. This simple filter was ≥ 1 ICD-9 codes for the phenotype of interest. All participants who passed this filter consisted of the “filter positive set.” Those without any ICD-9 codes for the phenotype were assigned a probability of 0. Comparisons of the above approaches against gold standard labels were performed among the filter positive set.
For the comparison, the main ICD features used in MAP were defined in 1 of 2 ways: either a list of ICD-9 codes defined by domain experts or generated using the semi-automated process as part of MAP. Similarly, the main NLP feature was defined either as a list created by domain experts or using the mapping steps described above. The optional denoising step in the PheNorm algorithm was not applied because it requires additional candidate features beyond the main ICD and NLP features, thereby limiting its ability to scale up for application to the PheWAS.
We evaluated the overall performance of the main ICD, main NLP, PheNorm, and MAP against the gold standard labels for the 16 phenotypes using the area under the receiver operating characteristic curve (AUC). In addition, we compared the MAP classification rule which automatically provides an estimated threshold to classify participants in phenotype yes/no categories. The performance of the MAP-based binary yes/no classification was compared to phenotypes defined using ≥ 2 ICD-9 codes. We reported the positive predictive value (PPV) or precision, negative predictive value (NPV), and F-score against the 16 phenotypes with gold standard labels.
MAP for genetic association studies
Using genetics and EHR data from Partners Biobank, we performed a genetic association study between a previously published LDL-C GRS34,35 and the disease phenotype of hyperlipidemia, which was defined either by using MAP predicted probabilities or based on having ≥ 2 ICD-9 codes for hyperlipidemia (ICD-9 272.x) and corresponding to the phecode 272.1.1,36 The association analyses were performed by fitting a logistic regression model regressing the MAP probabilities or ICD-9 based binary status for hyperlipidemia against the LDL-C GRS, adjusted for age, gender, and self-reported race.
MAP for PheWAS
In MVP, we performed a PheWAS for the IL6R variant, rs2228145. Individuals with the IL6R variant have profiles similar to individuals on IL6R antagonists, tocilizumab and sarilumab; both have been indicated for the treatment of rheumatoid arthritis (RA), and tocilizumab for giant cell arteritis (GCA). A recently published standard PheWAS based on ICD codes using MVP data found 22 phenotypes significantly associated with IL6R at the false discovery rate of 0.05.33 Although this single nucleotide polymorphism (SNP) was found to be associated with vascular and cardiac diseases, its associations with RA and GCA were not statistically significant. In this analysis, we tested whether MAP, when compared to the standard PheWAS method, improved the power to detect these expected associations.
For each phecode, we predicted the presence of the phenotype based on either the MAP algorithm or the ICD-9 code. Since MAP provided predicted probabilities of having the phenotype, we performed PheWAS directly relating the predicted probabilities from MAP to the genetic variant by fitting a quasi-binomial model. For both PheWAS methods, we adjusted for age, gender, and self-reported race—the robust sandwich estimator37 for the variance. We performed PheWAS on 1606 phenotypes, defined as the phecodes with prevalence > 0.1% in MVP.
RESULTS
The performance of MAP against existing approaches for phenotyping across 16 phenotypes
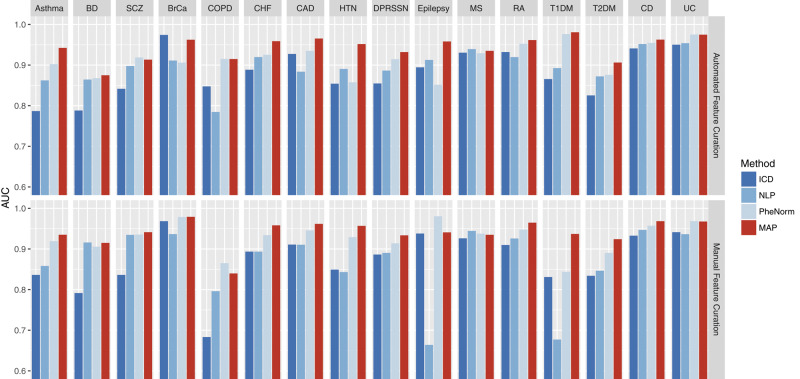
The ICD-9 and NLP concepts selected using the proposed semi-automated method as part of MAP yielded phenotype algorithms with similar AUCs to those developed using features selected by domain experts (Figure 1). The average AUC of the 16 MAP algorithms developed using ICD-9 + NLP features extracted using the MAP automated feature curation was 0.943, compared to 0.941 for algorithms developed from manual feature curation (Supplementary Table 1). The average AUCs of the individual ICD and NLP features created by the semi-automated process were 0.881 and 0.896—slightly higher than that of those curated manually (0.873 and 0.870, respectively). We focused on discussing the results using ICD-9 and NLP concepts selected by the proposed semi-automated method in the following sections.
Figure 1.
Top panel: Comparison of AUCs with gold standard labels for ICD-9 count, NLP, PheNorm, and MAP for 16 disease phenotypes using the MAP automated feature curation. Bottom panel: Comparison of AUCs for the 16 phenotypes features manually curated by domain experts.
Across the 16 phenotypes with gold standard labels, MAP classified the phenotypes with higher accuracy than ICD-9 or NLP in 13 out of the 16 cases (Figure 1, top panel). Also, the MAP algorithm outperformed PheNorm algorithm in most phenotypes and the most significant improvements in AUC were observed in Epilepsy and HTN (Supplementary Table 1). Compared to the ICD-9 feature, NLP feature, and PheNorm, the MAP algorithm improved the AUC significantly by about 0.062 (P = ), 0.047 (P = 2.91), and 0.027 (P = ) on average, respectively (Supplementary Table 1).
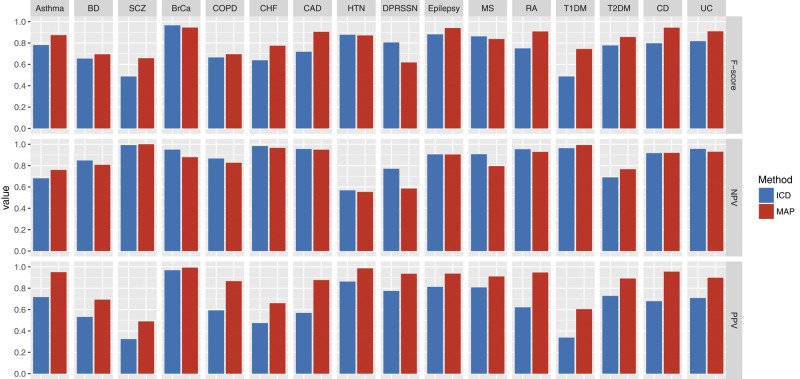
The MAP-based binary yes/no classification also had higher accuracy than phenotypes defined by using ≥ 2 ICD-9 codes when evaluated against a gold standard. Across the 16 phenotypes, MAP classified all phenotypes with a higher PPV (precision) than ICD-9 codes while maintaining a comparable NPV (Figure 2). For instance, the PPVs for the MAP and ICD-9 code were 0.947 versus 0.621 for RA and 0.876 versus 0.569 for CAD (Supplementary Table 2). Accounting for the trade-off between sensitivity (recall) and PPV (precision), the MAP classification attained a higher F-score than the simple ICD-9 rule across most phenotypes resulting in an average difference in F-score of 0.076 (P= ). In addition, MAP had larger AUCs (AUC here is for binary classifier) than ICD-9 codes across 16 phenotypes with an average difference of 0.136 (P = ). Similarly, MAP had better performance than binary classifier defined by NLP features (Supplementary Table 2).
Figure 2.
Performance of phenotype classification using MAP compared to ICD-9 codes for 16 phenotypes with gold-standard labels. (F-score, negative predictive value [NPV], positive predictive value [PPV, precision]).
Genetic association between LDL-C GRS and hyperlipidemia in the Partners Biobank
Among Partners Biobank participants, a total of 8226 (46.2%) out of 17 805 participants had ≥ 2 ICD-9 codes for hyperlipidemia. The estimated odds ratio (OR) between the LDL-C GRS and hyperlipidemia was 1.007 (P = .015) when hyperlipidemia was defined using the standard ICD-9 code thresholding approach. In contrast, the estimated OR was 1.010 (P = .0001) when the hyperlipidemia phenotype was defined using MAP, suggesting that MAP had improved power for association test compared to ICD-9 code.
PheWAS using VA MVP data
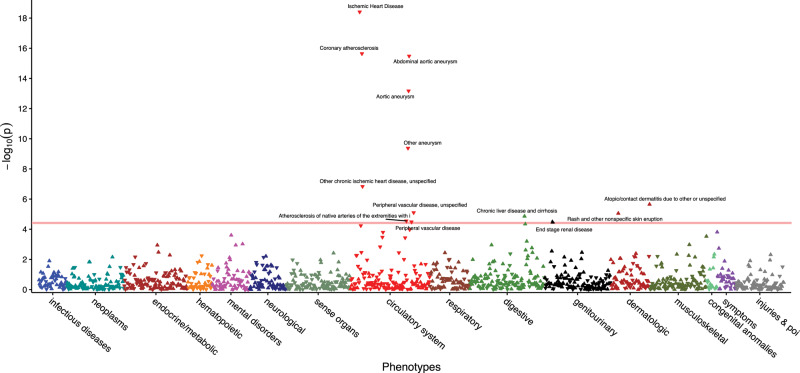
In MVP, we performed both MAP-based and ICD-9 based PheWAS of IL6R rs2228145. The minor allele frequency for IL6R rs2228145 (risk allele: A; Asp358Ala) was 0.35. The PheWAS results based on the MVP data are shown in Figure 3. Detailed results for the association pairs that attained significant P values after Bonferroni correction based on either MAP or ≥ 2 ICD-9 codes are shown in Supplementary Table 3. Compared to the standard ICD-9 code-based PheWAS, MAP yields a similar number of significant associations when using Bonferroni correction, 13 for MAP and 12 for ICD. MAP, using the predicted probability as the outcome, generally detected stronger associations with odds ratios in larger magnitude and smaller P values when compared to the results based on the standard PheWAS. For example, the estimated OR between IL6R SNP and ischemic heart disease was 0.945 (P = ) for MAP and 0.955 (P = for ICD. Among the 17 phenotypes that were significantly associated with IL6R by either MAP or standard PheWAS, the average negative log10 P value was 7.40 for MAP and 6.78 for ICD while the average absolute log odds ratio was estimated as 0.067 by MAP but 0.064 by ICD.
Figure 3.
PheWAS results using MAP-defined phenotypes for the IL6R SNP. Phenotypes significantly associated with IL6R after Bonferroni correction are annotated.
The IL6R antagonists were approved for treating rheumatoid arthritis (RA), giant cell arteritis (GCA), and systemic juvenile idiopathic arthritis (sJIA), also known as juvenile rheumatoid arthritis (JRA). However, the associations between IL6R and these 3 phenotypes were not detected in the VA MVP PheWAS due to a combination of low prevalence and low accuracy of the ICD codes.33 The IL6R PheWAS in MVP using ICD codes to define phenotypes found the following associations: GCA 0.926 (P = .279), RA 0.984 (P = .402), and JRA 1.082 (P = .684). Using MAP-based phenotype definitions, the following associations were observed with IL6R: GCA 0.883 (P = .082), RA OR 0.986 (P = .494), and JRA OR 1.152 (P = .509) (Supplementary Table 4). Defining phenotypes with MAP showed a stronger association between IL6R and reduced risk of GCA whereby the magnitude of the reduced risk was higher with MAP as well as the level of significance. For RA and JRA, neither MAP nor ICD detected associations with IL6R. In the VA population, there is a low prevalence of both conditions (particularly for JRA) and low accuracy of the ICD codes.
We also validated the association between IL6R and GCA, RA and JRA with data from the Partners Biobank. Using the ICD-based definition for phenotypes the associations with IL6R found the following: GCA OR 0.870 (P = .320), RA OR 0.897 (P = .008), and JRA OR 0.745 (P = .050). Using MAP to define the phenotypes, the estimated OR was 0.853 (P = .269) for GCA, 0.888 (P = .007) for RA, and 0.721 (P = .041) for JRA (Supplementary Table 4). These results again suggest improved power in detecting associations using the MAP approach.
DISCUSSION
In summary, MAP provided 3 major advancements for high-throughput phenotyping. First, MAP provides a systematic automated approach for integrating narrative information using NLP with ICD codes for use in a high-throughput phenotype algorithm pipeline. Second, MAP automates selection of the NLP concepts for a given phenotype, which is traditionally a labor-intensive manual process. Third, built into MAP is the ability to assign a threshold to provide a binary yes/no classification for each phenotype. This was accomplished by using the existing EHR data, thereby bypassing another traditionally rate-limiting step—that is, medical record review—to determine the threshold probability above which a participant is considered to have the phenotype. This high degree of automation allows the integration of more data which had been extracted using NLP into the algorithms, resulting in improvements in accuracy while maintaining the scalability and enabling large-scale phenotype screens such as the PheWAS. The improved power and scalability will also enable more efficient and less biased epidemiological studies since MAP can be used to accurately annotate both the main phenotype of interest and other risk factors or covariates. In addition, the scalability of MAP is enhanced by its efficiency in computation time. For the MVP cohort, it took about 1 minute per phenotype to run MAP utilizing 1 core on an Intel Xeon 5650 6-Core CPU (2.66 GHz) and it took NILE about 1 second to process 1000 notes that have at least 500 characters using the dictionary of 43 291 terms and 12 621 CUIs.
Similar to an existing high-throughput phenotyping approach such as PheNorm26 and others,24 MAP can rank order participants according to the likelihood of having the phenotype. However, MAP also provides a predicted probability of having the phenotype as well as an automated method to determine a threshold for each phenotype, facilitating classification into the binary phenotype yes/no categories. This ability to define a phenotype in 2 ways provides options for investigators depending on the study question. Using a probability of phenotype rather than a binary yes/no classification allows investigators to consider a phenotype as a continuous trait. This characteristic improved power to detect associations38 compared to the standard PheWAS approach, which uses a binary trait, with higher effect sizes and smaller P values. From our study using Partners Biobank, using MAP-defined hyperlipidemia yielded much more significant results than that defined by ICD code, regarding the association between the LDL-C GRS and hyperlipidemia. This further confirms that using MAP-defined phenotypes can improve the statistical power of downstream association studies as well as produce more accurate estimates of the effect sizes compared to ICD-based analyses.
The option of classifying participants into yes/no binary phenotype categories is also a step not available in PheNorm or other existing unsupervised algorithms. The ability to generate a data-driven threshold that is adaptive to the specific phenotype and the health care system is also a desirable feature. Existing methods largely use a common threshold (eg, 1 or 2 for ICD codes) but may result in poor performance for specific phenotypes or poor portability due to heterogeneity in health care systems.10 Prevalence estimates using MAP, ICD codes, and chart review are provided in Supplementary Table 2, which suggests that the MAP estimates are more consistent with those based on chart reviews. Throughout our experiments, we assigned a probability of 0 for patients in the filter negative set and evaluation on the filter positive set. The filter defined as having at least 1 ICD code generally has a high NPV. For example, in Partners Biobank, the NPV for having no ICD code attained an average of 99% across the 16 phenotypes—10 of which reached 100%. In addition, the average NPV across 16 phenotypes for all patients (including both filter negative and filter positive patients) was 0.994 for both MAP and ICD-9 code (Supplementary Table 5). These suggest that assigning a probability of 0 for filter negative patients is safe and reasonable. However, alternative filters may perform better, and our MAP approach is not restricted to any specific filter. For undercoded disease phenotypes such as suicide ideation, filters based on both NLP and ICD may attain higher accuracy.
A key part of MAP is the use of automated steps to identify the relevant ICD or NLP features for any given phenotype algorithm. The algorithms developed using ICD and NLP features selected using the MAP process had comparable or slightly better AUCs than manually curated features selected by domain experts across the 16 phenotypes. Additionally, the experiment shows that MAP was more robust than PheNorm. MAP uses model averaging over 3 types of features (ICD, NLP, and ICD+NLP) and 2 types of distributions (Poisson and log-normal) for algorithm development; PheNorm uses only the normal distribution and relies on majority voting. The additional models used by MAP improves the robustness of the algorithm. Specifically, to see the impacts of each single model and health care utilization on the performance of the MAP algorithm, we compared the performance of our proposed MAP algorithm to several simplified versions of MAP algorithm by removing some of the components for phenotyping the 16 diseases. From the results (Supplementary Tables 1 and 2), we found that the performance of MAP algorithm decreased when some of the components were excluded. To aggregate the results from all 6 models, MAP currently uses a simple model average of the predicted probabilities, in part due to the difficulty in ascertaining the performance of mixture models in the absence of labels. Data adaptive weighting can potentially be used to further improve the MAP performance. The current implementation of MAP counts ICD-9 codes with ICD-10 codes mapped to ICD-9 based on GEMS. Ambiguous or uncertain mappings are not currently excluded. An alternative approach to incorporating ICD-10 codes is to use the recently updated phecode mapping that includes ICD-10 codes.39
It is worth noting that the denoising step of the original PheNorm was not used in this study. The denoising step can potentially improve the accuracy of PheNorm when the main ICD and NLP features are not sufficiently predictive (eg, in the cases of asthma with automated features and type I diabetes mellitus with manual features). In comparison, MAP had a more robust performance and can attain reasonable accuracy in the absence of additional features and is thus highly scalable. However, it is plausible that incorporating additional features may further improve the accuracy of MAP for certain phenotypes which warrants further research.
Compared to ICD-based analyses, the improved accuracy of MAP translated into improved power in both association studies and a PheWAS using real-world cohort data. From the genetic association study example between the LDL-C GRS and hyperlipidemia in the Partners Biobank (n = 17 805), we observed that improving the accuracy of phenotype definitions is particularly important when the sample size is relatively small. The association between LDL-C GRS and hyperlipidemia was much more significant when the phenotype is defined by MAP compared to that defined by ICD-9 codes alone. In MVP and Partners Biobank, MAP provided improved power to detect associations between IL6R and uncommon phenotypes where the accuracy of the ICD code was relatively low.
CONCLUSION
Using a relatively large number of phenotypes with gold standard labels for validation, we demonstrated that the proposed MAP algorithm achieves more efficient, robust, and accurate phenotyping compared to existing approaches. A distinct advantage to MAP is the process of providing automated selection of both ICD and NLP features and integration of multiple feature types and distributions. We validated the MAP approach in 2 independent biobanks, which demonstrated that MAP annotated phenotypes are comparable to those developed using manual approaches, can replicate known associations, and improves power in smaller data sets. Finally, MAP provides 2 forms of phenotype data for studies: 1) in a traditional phenotype yes/no format or 2) as a probability of a phenotype. The MAP high-throughput phenotyping approach integrating ICD-9 and key NLP concepts has direct applications in large-scale association studies such as PheWAS in biorepositories linked with EHR data. This promising approach can also improve the resolution, particularly among less common or poorly coded conditions, to study the relationships across multiple phenotypes in 1 study.
FUNDING
This work was supported in part by the US National Institutes of Health grants P30-AR072577, U54-HG007963, U01-HG008685 and RO1-HG009174; National Natural Science Foundation of China grant 11801301; and National Key R&D Program of China grant 2018YFC0910404.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to: conception and design, acquisition, analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, and final approval of the version to be published.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement
None declared.
Supplementary Material
REFERENCES
- 1. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010; 269: 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benesch C, Witter DM Jr., Wilder AL, Duncan PW, Samsa GP, Matchar DB.. Inaccuracy of the International Classification of Diseases (ICD-9-CM) in identifying the diagnosis of ischemic cerebrovascular disease. Neurology 1997; 493: 660–4. [DOI] [PubMed] [Google Scholar]
- 3. Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF.. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care 2005; 435: 480–5. [DOI] [PubMed] [Google Scholar]
- 4. White RH, Garcia M, Sadeghi B, et al. Evaluation of the predictive value of ICD-9-CM coded administrative data for venous thromboembolism in the United States. Thromb Res 2010; 1261: 61–7. [DOI] [PubMed] [Google Scholar]
- 5. Zhan C, Battles J, Chiang YP, Hunt D.. The validity of ICD-9-CM codes in identifying postoperative deep vein thrombosis and pulmonary embolism. Jt Comm J Qual Patient Saf 2007; 336: 326–31. [DOI] [PubMed] [Google Scholar]
- 6.CONSIDER-A COMPUTER PROGRAM FOR MEDICAL INSTRUCTION. N Y State J Med 1969. MED SOC STATE OF NY 420 LAKEVILLE RD PO BOX 5404, LAKE SUCCESS, NY 11042.
- 7. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of electronic health record oriented phenotyping algorithms. AMIA Annu Symp Proc 2011; 2011: 274–83. [PMC free article] [PubMed] [Google Scholar]
- 9. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013; 20 (e1): e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010; 628: 1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn's disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach . Inflamm Bowel Dis 2013; 197: 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia Z, Secor E, Chibnik LB, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS One 2013; 811: e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castro VM, Minnier J, Murphy SN, et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am J Psychiatry 2015 1724: 363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu S, Kumamaru KK, George E, et al. Classification of CT pulmonary angiography reports by presence, chronicity, and location of pulmonary embolism with natural language processing. J Biomed Inform 2014; 52: 386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS One 2015; 108: e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao KP, Cai T, Savova GK, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. BMJ 2015; 350: h1885.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castro V, Shen Y, Yu S, et al. Identification of subjects with polycystic ovary syndrome using electronic health records. Reprod Biol Endocrinol 2015; 13: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castro VM, Dligach D, Finan S, et al. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology 2017; 882: 164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 236: 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Carroll RJ, Hinz ER, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc 2013; 20 (e2): e253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu S, Liao KP, Shaw SY, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc 2015; 225: 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J Am Med Inform Assoc 2017; 24 (e1): e143–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiu PH, Hripcsak G.. EHR-based phenotyping: bulk learning and evaluation. J Biomed Inform 2017; 70: 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 236: 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Halpern Y, Horng S, Choi Y, Sontag D.. Electronic medical record phenotyping using the anchor and learn framework. J Am Med Inform Assoc 2016; 234: 731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu S, Ma Y, Gronsbell J, et al. Enabling phenotypic big data with PheNorm. J Am Med Inform Assoc 2018; 251: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013; 3112: 1102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CMS.gov. 2018 ICD-10 CM and GEMS. 2018. https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.html. Last accessed on July 25, 2017.
- 29. Yu S, Cai T. A short introduction to NILE. CoRR arXiv: 1311.6063; 2013.
- 30. Karlson EW, Boutin NT, Hoffnagle AG, Allen NL.. Building the Partners Healthcare Biobank at Partners personalized medicine: informed consent, return of research results, recruitment lessons, and operational considerations. J Pers Med 2016; 61: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gainer VS, Cagan A, Castro VM, et al. The biobank portal for Partners Personalized Medicine: a query tool for working with consented biobank samples, genotypes, and phenotypes using i2b2. J Pers Med 2016; 61: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70: 214–23. [DOI] [PubMed] [Google Scholar]
- 33. Cai T, Zhang Y, Ho YL, et al. Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy: a phenome-wide association study. JAMA Cardiol 2018; 39: 849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010; 4667307: 707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao KP, Diogo D, Cui J, et al. Association between low density lipoprotein and rheumatoid arthritis genetic factors with low density lipoprotein levels in rheumatoid arthritis and non-rheumatoid arthritis controls. Ann Rheum Dis 2014; 736: 1170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013; 1510: 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Halekoh U, Højsgaard S, Yan J.. The R package GEEPACK for generalized estimating equations. J Stat Softw 2006; 152: 1–11. [Google Scholar]
- 38. Sinnott JA, Dai W, Liao KP, et al. Improving the power of genetic association tests with imperfect phenotype derived from electronic medical records. Hum Genet 2014; 13311: 1369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu P, Gifford A, Meng X, et al. Developing and evaluating mappings of ICD-10 and ICD-10-CM codes to phecodes. BioRxiv 2018; 462077. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.