Fig. 1.
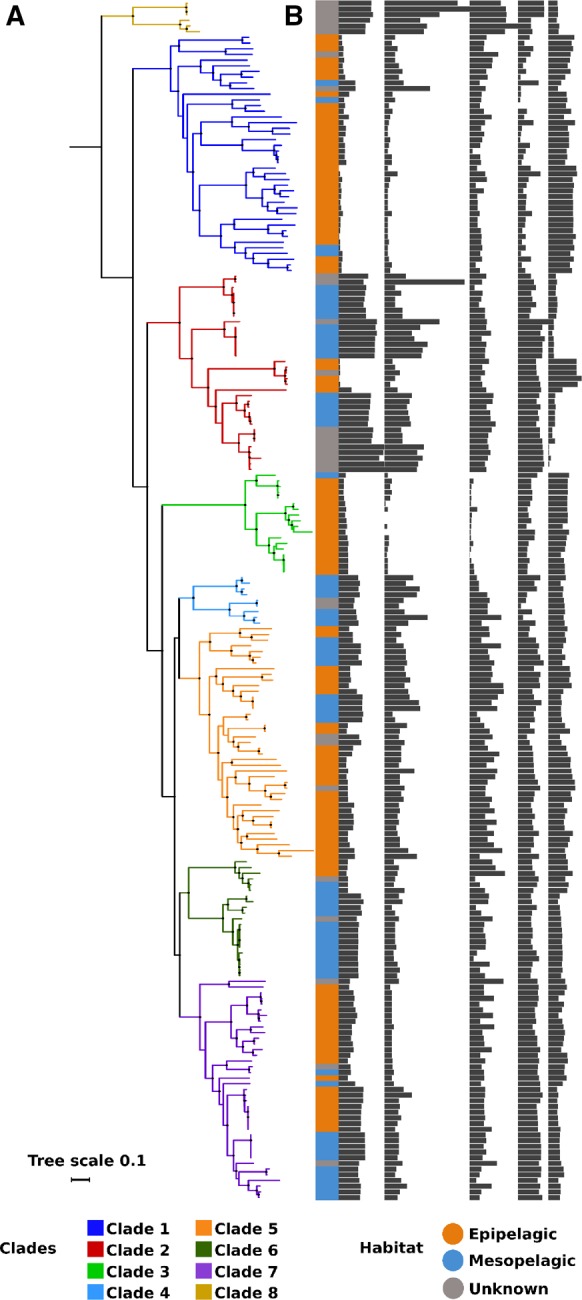
—Representation of phylogeny, habitat classification, and genomic features of Marinimicrobia. (a) Maximum likelihood phylogenetic tree of the 211 genomes constructed using amino acid sequences of 120 highly conserved marker genes. (b) Habitat classification based on Getz et al. (2018) and genomic features of Marinimicrobia genomes. Abbreviations: H, Habitat; GC, % GC content (range, 27–55%); IGR, median intergenic region length (range, 7–78 nucleotides); EGS, estimated genome size (range 1–4.4 Mb); N-ARSC (range, 0.3–0.34); C-ARSC (range, 3–3.2). Black points on branches represent support values >0.95.
