Figure 1.
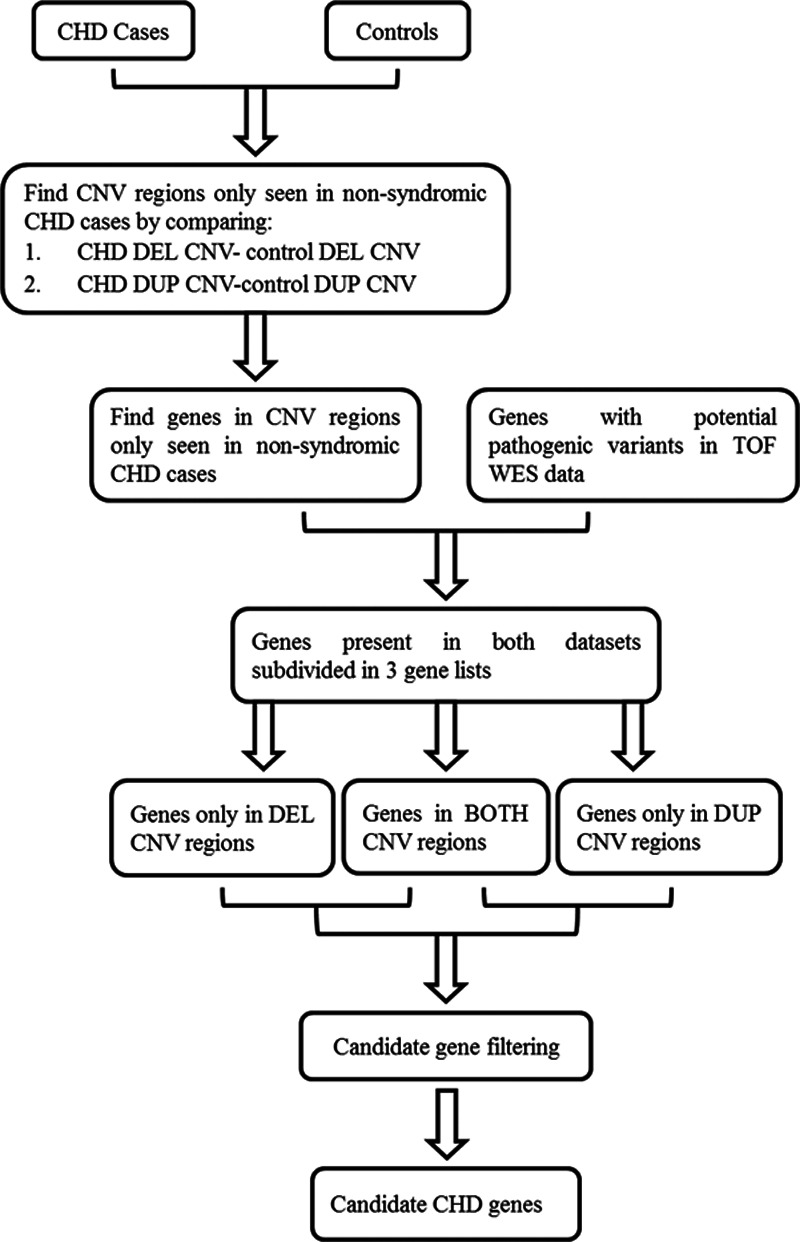
Overall methodology. Flowchart showing the methodology used to identify novel genetic loci for nonsyndromic congenital heart disease (CHD) cases. Potential pathogenic variants were novel or rare single nucleotide variants (either absent from ExAC or with frequency of <0.01). Candidate genes identified at the end of the workflow were subsequently analyzed for ohnolog status. BOTH indicates deleted/duplicated; CNV, copy number variant; DEL, deletion; DUP, duplication; TOF, Tetralogy of Fallot; and WES, whole-exome sequencing.
