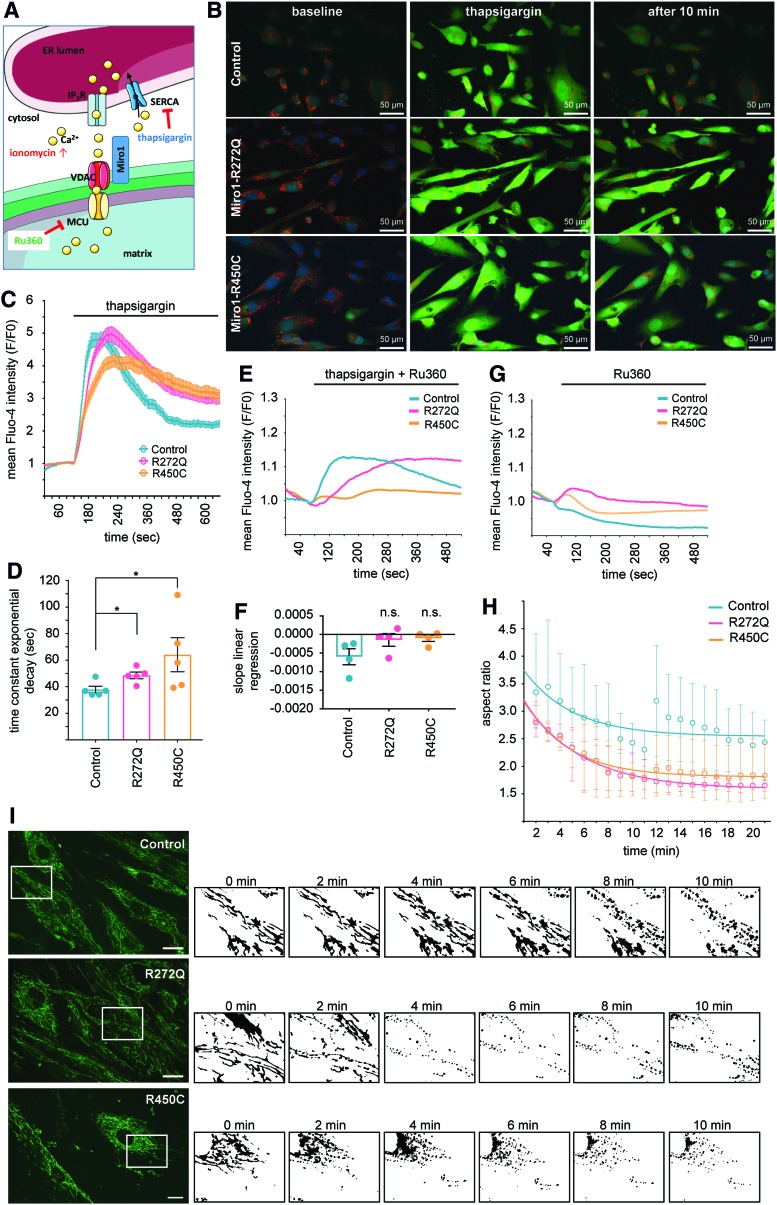
FIG. 2.
Reduced calcium buffering capacity in Miro1-mutant fibroblasts. (A) Overview of treatments with thapsigargin, ionomycin, and Ru360 for calcium imaging. Thapsigargin inhibits calcium uptake of the ER by the SERCA pumps, which leads to increase of cytosolic calcium levels by depletion of ER calcium stores. Ionomycin increases the levels of cytosolic calcium by activation of G-protein-coupled receptors in the plasma membrane. Ru360 is an inhibitor of the MCU. (B) Immortalized fibroblasts were loaded with Fluo4-AM (green) for live cell imaging. Cells were imaged under baseline condition for 2 min. After addition of 1 μM thapsigargin, imaging was continued for 10 min with a 2 s interval. Images were acquired using a 25 × objective; scale bars indicate 50 μm. (C) Quantification of calcium levels upon thapsigargin treatment from (B). Mean intensity of Fluo4-AM signal was indicated as (F/F0). Data indicated as mean ± SEM, n = 5, with 8–20 cells per cell line per experiment. (D) Time constant of exponential decay calculated from calcium response curves of (C). Data indicated as mean ± SEM. Significance calculated using the Mann–Whitney test (n = 5). (E) Immortalized fibroblasts were stained with Fluo4-AM for live cell imaging. After 1 min of baseline recording, cells were treated with a combination of 10 μM Ru360 and 1 μM thapsigargin, and imaging was continued for 9 min, using a 25 × objective (n = 3–4). Data indicated as mean. (F) Slope of linear regression of calcium response curves from (E). Data indicated as mean ± SEM. Significance calculated using the Mann–Whitney test (n = 3–4). (G) Immortalized fibroblasts were stained with Fluo4-AM and treated with 10 μM Ru360 during live cell imaging. Data indicated as mean (n = 3). (H) Microscopy images from (I) were analyzed for aspect ratios of the mitochondrial network over time using the ImageJ (n = 3–5). (I) Immortalized fibroblasts were stained with MitoTracker® Green FM and treated with 20 μM ionomycin during 20 min of live cell imaging, using a 63 × objective. Scale bars indicate 20 μm. Representative images of the mitochondrial network in control fibroblasts, Miro1-R272Q, and R450C fibroblasts. Mitochondrial masks generated with the ImageJ for the analysis of mitochondrial morphology were displayed at different time points for all cell lines. The white boxes in the microscopy images on the left highlight the sections depicted on the right, showing details of the mitochondrial network at different time points (binary images). Significance for all data calculated by the Mann–Whitney test. *p ≤ 0.05. ER, endoplasmic reticulum; MCU, mitochondrial calcium uniporter; SEM, standard error of the mean. Color images are available online.