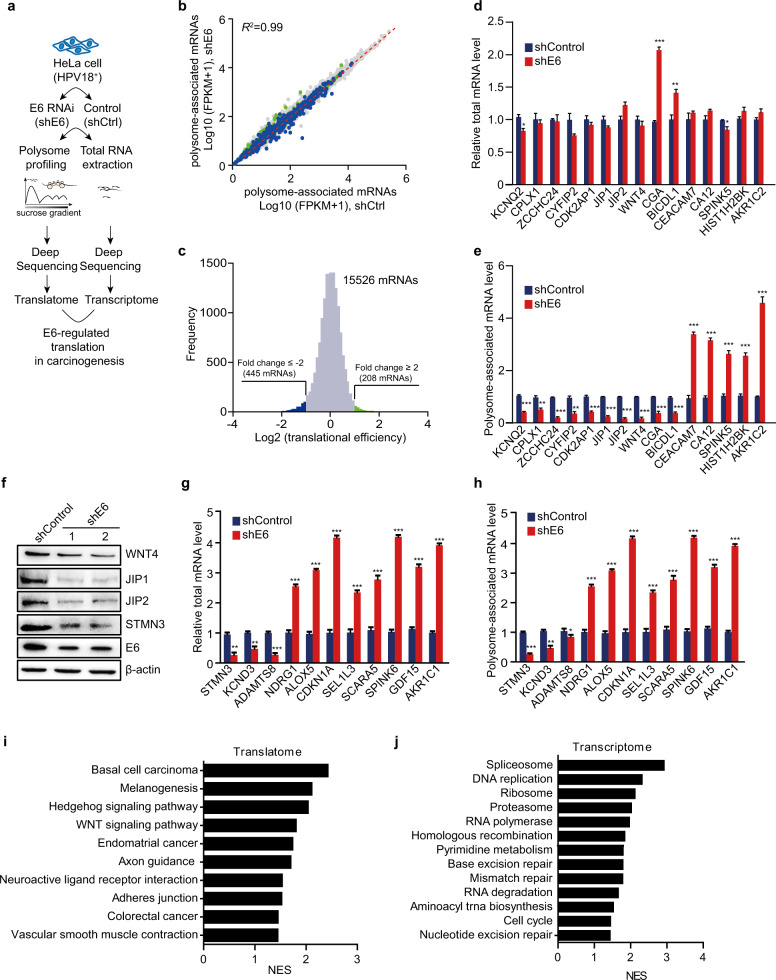
Fig. 1.
E6 of HPV18 affects mRNA translation in HeLa cells. a Schematic of the protocol for identifying E6-regulated mRNA translation at the transcriptome level. b Counts of polysome-associated mRNAs (fragments per kilobase per million mapped fragments, FPKMs) from E6-silenced (shE6) and control (shControl or shCtrl) cells. mRNAs with over a two-fold decrease or increase are indicated in blue and green, respectively. c Distribution of changes in translation efficiency for shControl and shE6 cells. mRNAs with decreased (fold change ≤ −2) or increased (fold change ≥ 2) translation efficiencies are indicated as above. d, e Some mRNAs with inconsistent changes between the transcription level and translation level were further confirmed by qRT-PCR. The transcription levels (d) and translation levels (e) were measured using total RNAs or polysome-associated mRNAs. The data represent means ± SEM from at least three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001). f The protein levels of the indicated genes in shE6 and shControl HeLa cells were determined by western blot. g, h The transcription levels (g) and translation levels (h) of the indicated mRNAs with consistent changes were measured by qRT-PCR as described in d and e. The data represent means ± SEM from at least three independent experiments (*p < 0.05, **p < 0.01, ***p < 0.001). i, j Top pathways (E6-activated pathways) enriched in shControl HeLa cells from GSEA of RNA-Seq data for polysome-associated mRNAs (i) or the transcriptome (j)