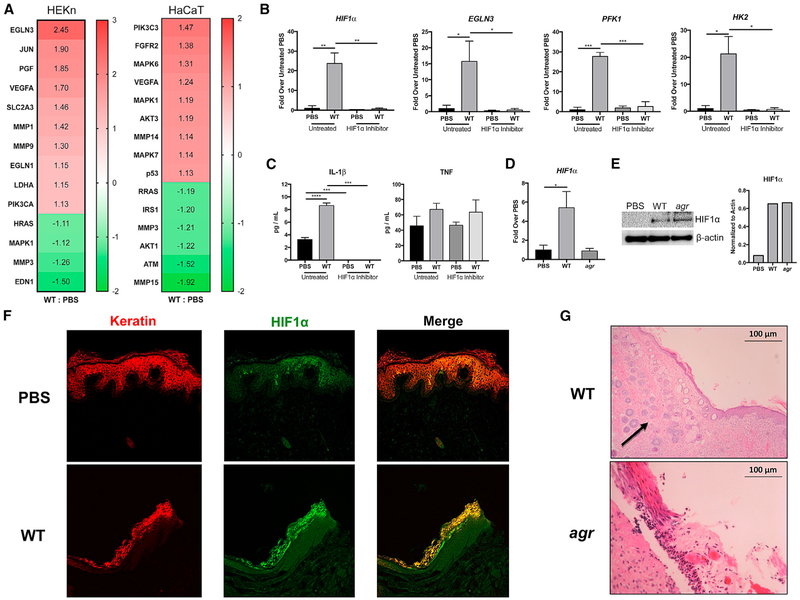
Figure 2. Staphylococcal Induction of HIF1α in Keratinocytes.
(A) An Ingenuity Pathway Analysis (IPA) was performed on RNA-seq data obtained from both human keratinocytes in primary culture (HEKn) and HaCaTs exposed to S. aureus (WT) USA300 LAC for 1 hr (MOI, 100:1). Heatmap shows the relative activation of genes responsive to HIF1α.
(B and C) qRT-PCR (B) and ELISAs (C) were performed on keratinocytes in primary culture exposed to PBS or WT S. aureus, with and without a small-molecule HIF1α inhibitor.
(D and E) qRT-PCR (D) and immunoblot (E) of HIF1α in primary keratinocytes after 24 hr of WT or agr infection. Densitometry normalized to actin is included for comparison.
(F) Immunofluorescence staining of infected and control human skin grafts with α-keratin (red) and α-HIF1α (green) demonstrating co-localization (yellow) in the infected grafts.
(G) H&E staining of sections from infected skin grafts. Arrow indicates areas of neovascularization.
Representative data from at least two independent experiments are shown. For all graphs, each data point is the mean value ± SEM (n = 4). *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001, by one-way ANOVA.