Figure 5. The discovery of acetylation of K134 in MsNrtR.
(A) Use of Western blot to probe the acetylation of recombinant MsNrtR protein in both E. coli and M. smegmatis. The two forms of recombinant NrtR protein were purified from E. coli BL21 and M. smegmatis, and analyzed by western blotting using both anti-acetyl-lysine antibody (α-Acetyl) and poly anti-MsNrtR rabbit serum. The bigger version of MsNrtR is produced by the pET28 expression plasmid in E. coli, whose N-terminus is fused to the 6xHis-containing tag of 23 residues (Supplementary file 1). By contrast, the smaller version of MsNrtR is generated by pMV261 in M. smegmatis, which is only tagged with C-terminal 6xHis. The altered molecular mass (~2 kDa) is the reason why the migration rate of protein electrophoresis differs slightly for the two MsNrtR versions. A representative result is given from three independent trials. (B) The discovery of a unique Lys134 acetylation site in MsNrtR. A LC/MS spectrum reveals that a charged peptide (LVAkLSYTNIGFALAPK) of MsNrtR bears an acetylated lysine (K134Ac). The sequence depicted in the yellow box illustrates the K134 site of acetylation in the context of the modeled structure of MsNrtR-DNA. (C) The mutation of K134A results in reduced acetylation of MsNrtR in M. smegmatis MC2 155 (Magni et al., 2004). A representative result from three independent experiments is given.
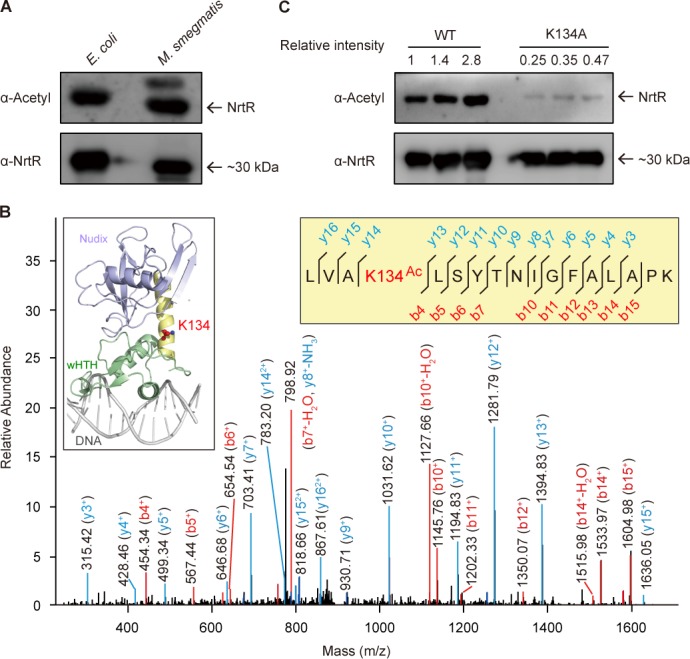
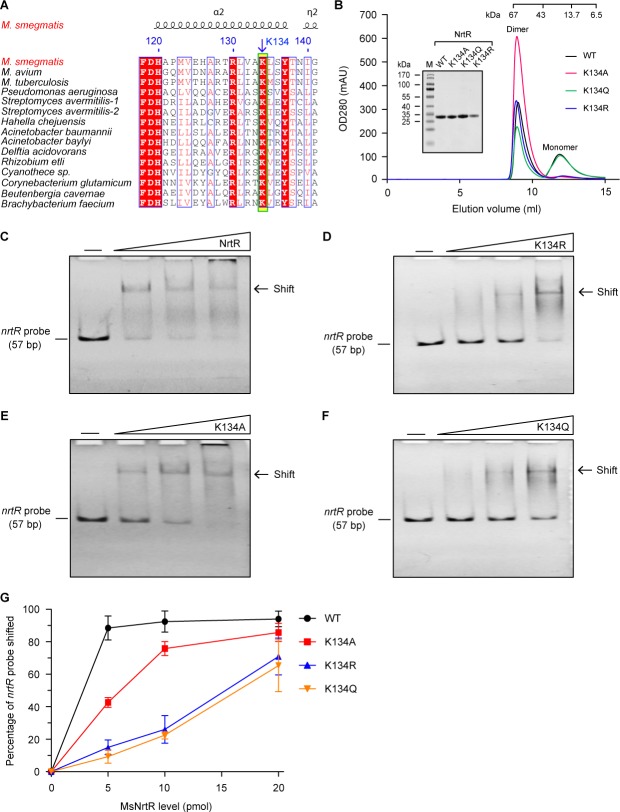
Figure 5—figure supplement 1. Dependence on K134 acetylation in the binding of MsNrtR to a cognate DNA target.
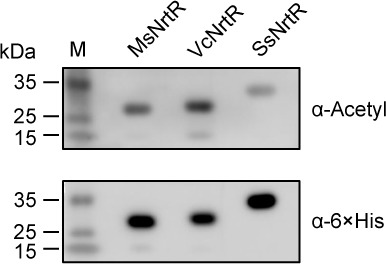
Figure 5—figure supplement 2. Acetylation is ubiquitous in three bacterial NrtR proteins.