Scheme 1.
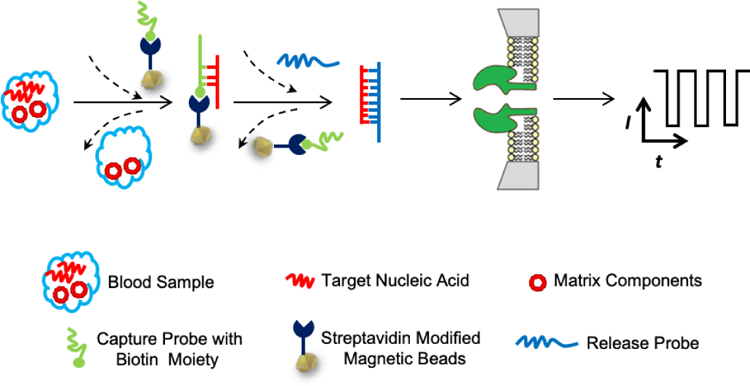
Schematic representation of the principle of displacement chemistry-based nanopore analysis of nucleic acids in complicated matrices. The separation of the target nucleic acid from the sample matrix components and its sequence analysis are achieved by using two ssDNA probes. The capture probe, which is immobilized on the magnetic beads, has sequences partially complementary to the target nucleic acid, while the release probe has a much stronger hybridization affinity to the target nucleic acid than the capture probe does. After addition of the capture probe to the blood or serum sample, the target nucleic acid molecules in the blood and the capture probe will form sticky end double-stranded nucleic acid molecules, and are immobilized on the magnetic beads, thus being separated from other matrix components. After removing the blood followed by addition of the release probe, the target nucleic acid forms more stable double-stranded nucleic acid with the release probe than with the capture probe, thus being pulled off from the magnetic beads. The produced release probe-nucleic acid hybridization complexes can then be transferred to the nanopore sensor for determining the sequence of the target nucleic acid.
