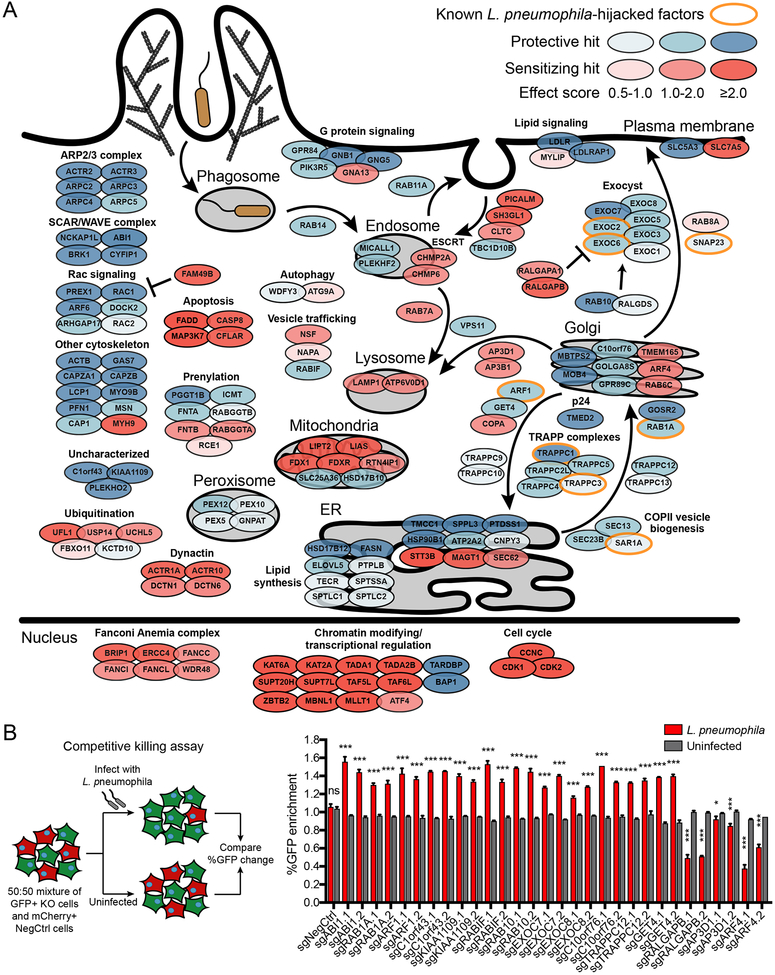
Figure 2. Summary of modifiers of L. pneumophila pathogenesis from CRISPR screens.
(A) Schematic of selected hits in cellular compartments or processes. Genes are color-coded by effect size. Previously known host factors hijacked by L. pneumophila are outlined in orange. Complete batch retest screen results are in Table S3.
(B) Validation of selected hits using individual sgRNAs in a competitive killing assay. Knockout or negative control cells (GFP+) were cocultured with negative control cells (mCherry+) in equal numbers and either infected with L. pneumophila at MOI = 100 or left uninfected. The percentage of GFP+ cells was measured by Incucyte and the % GFP change at 48 h was compared between L. pneumophila-infected and uninfected populations (n = 3 technical replicates, two-tailed Student’s t test, ns = not significant, *P<0.05, ***P<0.001; error bars, s.d.). Data shown are representative of 2 independent experiments.
See also Figure S2.