Abstract
During polyketide and fatty acid biosynthesis, the growing acyl chain is attached to the acyl carrier protein via a thioester linkage. The acyl carrier protein interacts with other enzymes that perform chain elongation and chain modification on the bound acyl chain. Most type I polyketide synthases and fatty acid synthases contain only one acyl carrier protein. However, polyunsaturated fatty acid synthases from deep-sea bacteria contain anywhere from two to nine acyl carrier proteins. Recent studies have shown that this tandem acyl carrier protein feature is responsible for the unusually high fatty acid production rate of deep-sea bacteria. To investigate if a similar strategy can be used to increase the production rate of type I polyketide synthases, a 3×ACP domain was rationally designed and genetically installed in module 6 of 6-deoxyerythronolide B synthase, which is a prototypical type I modular polyketide synthase that naturally harbors just one acyl carrier protein. This modification resulted in an ~2.5-fold increase in the total amount of polyketide produced in vitro, demonstrating that installing a tandem acyl carrier domain in a type I polyketide synthase is an effective strategy for enhancing the rate of polyketide natural product biosynthesis.
Graphical Abstract
Polyketides are a class of secondary metabolites synthesized by plants, insects, fungi, and bacteria.1 The name “polyketide” refers to the multiple ketide groups (─CH2─ CO─) found in these natural compounds. In nature, polyketides confer competitive advantage to the host organism such as antimicrobial activity and pigmentation. Many polyketide natural products also exhibit useful pharmacological properties and some have been developed into successful therapeutics. FDA-approved polyketide drugs include erythromycin (antibiotic), streptomycin (antibiotic), and doxorubicin (anticancer). One of the greatest challenges in polyketide drug development is securing large quantities of the natural product needed to carry out preliminary studies and clinical trials. This is because many polyketides are produced at low levels by the native producer or they are produced by unculturable organisms. Therefore, a vast number of promising polyketides remain inaccessible for drug research. One possible solution to this problem is to engineer the polyketide synthase (PKS) enzymes responsible for polyketide biosynthesis in the native host to increase the product yield.
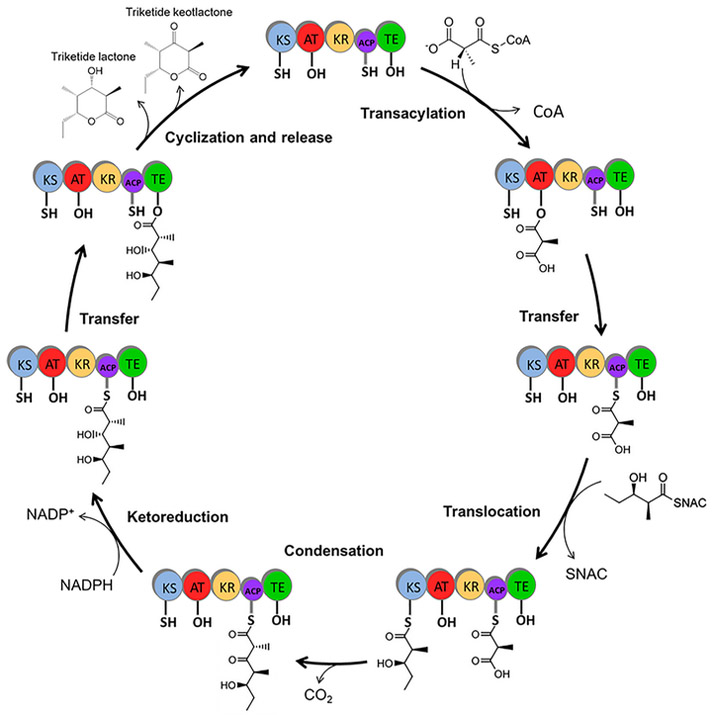
In this study, we sought to improve the product turnover of a modular PKS enzyme by incorporating additional acyl carrier proteins (ACPs). We used 6-deoxyerythronolide B synthase (DEBS), which is a type I modular PKS expressed by the soil bacterium Saccharopolyspora erythraea, as the model system. Three DEBS proteins work together in successive fashion to produce the macrolide ring of the antibiotic erythromycin (see Figure S1 in the Supporting Information).2 Each DEBS protein contains two modules, and each module contains a varying number of enzymatic domains that cooperatively perform one round of polyketide chain extension and optional reductive modification. The acyltransferase (AT) domain selects the extender unit, methylmalonyl-coenzyme A, from the cytoplasm. The ketosynthase (KS) domain catalyzes carbon–carbon bond formation between the extender unit and the growing polyketide chain. The ACP receives the extender unit from the AT domain and carries it to the KS domain, where it becomes covalently attached to the growing polyketide chain. Next, the ACP transfers the extended polyketide chain to the optional reductive domains (ketoreductase, enoylreductase, and dehydratase), where the newly formed β-keto group becomes modified to a hydroxyl group or a fully reduced carbon atom. Finally, the ACP passes the extended polyketide chain to the KS domain of the downstream module for subsequent round of chain extension and modification, or to the thioesterase (TE) domain for product release.3
Our protein engineering study was performed on module 6-TE which is a single-module PKS derived from module 6 of DEBS 3. Module 6-TE consists of KS, AT, KR, ACP, and TE domains (see Figure 1). We chose this PKS protein because (i) its small size allows for facile genetic manipulation and (ii) the presence of the TE domain makes direct product detection possible. DEBS module 6-TE has previously been shown to accept the non-natural substrate (2S,3R)-2-methyl-3-hydroxypentanoic-N-acetylcysteamine thioester and the methylmalonyl coenzyme A extender unit to form a mixture of triketide lactone (TKL) and triketide ketolactone (TKKL) product in a ratio of 1:1.8.4 The nonreduced product, TKKL, is thought to be formed when the TE domain acts on the ACP-bound linear triketide before the KR domain.
Figure 1.
Module 6-TE catalyzes chain elongation, β-keto group modification, and lactone formation on the model substrate (2S,3R)-2-methyl-3-hydroxypentanoic-N-acetylcysteamine thioester.
In nature, ACPs are found in all PKSs and fatty acid synthases.5 ACPs are small (80–110 amino acids), generally acidic (pI = 5–6) proteins with a four-helix bundle structural motif.6-10 ACPs have a conserved active site serine that is post-translationally modified with a 4′-phosphopantetheine prosthetic group to form the functional holo-ACP. This modification provides a long, flexible arm that contains a terminal thiol group to which the growing acyl chain is attached during polyketide and fatty acid biosynthesis. Typically, PKSs and fatty acid synthases contain a single ACP. But polyunsaturated fatty acid (PUFA) synthases from deep-sea bacteria universally contain multiple (2–9) ACPs that are arranged in series, collectively known as a “tandem ACP domain”. For example, PUFA synthase from Moritella marina, Shewanella japonica, and Schizochytrium sp. contains five, six, and nine consecutive ACPs, respectively (see Figure S2 in the Supporting Information).11-13 The tandem ACP domain of PUFA synthases are positioned between the malonyl-CoA transferase and the ketoreductase domains. This is in contrast to PKSs and mammalian fatty acid synthases whose single ACP domain is located downstream of the reducing domains.
Shen and colleagues studied the 6×ACP domain from Shewanella japonica and found that the six individual ACPs, which share 85%–96% sequence identity, are functionally equivalent.14 Inactivating one or more ACPs in this tandem ACP domain by mutating the active site serine to alanine resulted in proportionately decreased fatty acid production. This result indicates that the tandem ACP domain becomes the rate-limiting element of the fatty acid biosynthesis pathway when the number of ACPs is artificially reduced. Baerga-Ortiz and colleagues investigated the solution structure of the isolated 5×ACP domain from Photobacterium profundum, using small-angle X-ray scattering (SAXS), and derived an elongated beads-on-a-string model of the tandem ACP domain, indicating that there is little or no interaction between the individual ACPs in a tandem ACP system.15 These preliminary findings suggest that adding extra ACPs in a PKS module that natively contains only one ACP will increase its product turnover number. To test this idea, we have genetically inserted two extra ACPs to DEBS module 6-TE, thus creating module 6–3ACP-TE.
RESULTS AND DISCUSSION
Amino Acid Sequence Alignment of Natural Tandem ACPs and de Novo Design of an Artificial Tandem ACP.
Three variables must be considered in designing an artificial tandem ACP domain: how many ACPs to incorporate, the amino acid sequence of the individual ACPs, and the length and amino acid sequence of the linker regions that lie between the ACPs. To understand how nature constructs tandem ACPs, we analyzed the primary structure of naturally occurring tandem ACPs (see Figure S3 in the Supporting Information). PKSs that natively contain tandem ACPs have relatively small tandem ACP domains (consisting of two or three ACPs), whereas marine PUFA synthases typically have larger tandem ACP domains (consisting of as many as nine ACPs). The tandem ACP domain of PKSs consists of ACPs with varying degree of amino acid sequence homology (20%–95% sequence identity), while the tandem ACP domain of PUFA synthases is composed of ACPs having a much higher sequence homology (80%–100% sequence identity). The ACP-to-ACP linker region is highly variable in length and amino acid composition for both PKSs and PUFA synthases. We decided to create module 6–3ACP-TE by faithfully triplicating the amino acid sequence of the native ACP of module 6-TE. This ACP has moderate sequence homology (14%–28% sequence identity) to the ACPs in naturally occurring tandem ACP domains. The three ACPs were linked via a 20-residue linker, TPAVGAVPAVQAAPAREMTS, whose sequence was obtained directly from the region immediately upstream of ACP in module 6-TE. Complete amino acid sequence of module 6–3ACP-TE is shown in Figure 2.
Figure 2.
Amino acid sequence of DEBS module 6–3ACP-TE. [Legend: ACP, acyl carrier protein (purple); AT, acyltransferase (red); KR, ketoreductase (yellow); KS, ketosynthase (blue); and TE, thioesterase (green). Linker regions are shown in black.]
Comparison of Product Formation by Native and Engineered DEBS Module 6-TE.
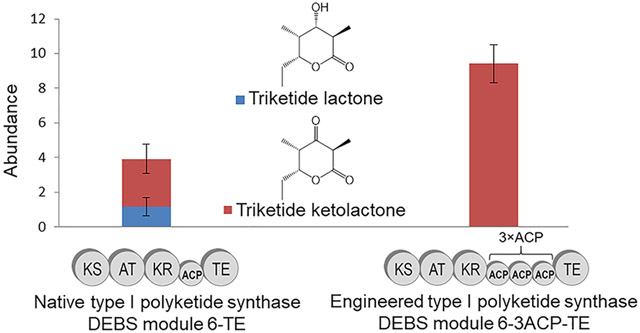
We produced the module 6-TE and module 6–3ACP-TE proteins in E. coli BAP1, which constitutively expresses the Bacillus subtilis Sfp (4′-phospho-pantetheinyl transferase) to promote in vivo conversion of apo- to holo-ACP.16-18 The purified proteins were also incubated with purified recombinant Sfp and co-enzyme A to further promote conversion to holo-ACP. The in vitro catalytic activity of module 6-TE and module 6–3ACP6-TE was confirmed using liquid chromatography–mass spectrometry (LC-MS), following an established protocol.19-23 The products, TKL and TKKL, could be readily identified in the mass spectrum (see Figures S5, S6, and S7 in the Supporting Information). A 7-h time-course study revealed that module 6-TE produced both TKL and TKKL, as reported previously,4 while module 6–3ACP-TE exclusively produced TKKL (see Figure 3). It is not clear why module 6–3ACP-TE did not produce any TKL. One possibility is that the individual ACPs in the tandem ACP domain are unable to interact with the KR due to crowding. Another possibility is that the ACP bound substrate gets hydrolyzed by the TE domain before interaction with the KR is established. Notwithstanding the absence of KR activity, the total amount of macrolactone formed by module 6–3ACP-TE is ~2.5 times greater than that formed by module 6-TE, and this result is consistent with previous findings on naturally occurring tandem ACP systems that the number of ACPs is proportional to product output. In order to confirm that the rate enhancement observed in module 6–3ACP-TE is due to the tandem ACP domain, and not from bypassing of the ketoreduction step, we performed the triketide formation assay with the native module 6-TE in the absence of NADPH. As expected, only TKKL was formed by module 6-TE when NADPH was not added (see Figures S8 and S9 in the Supporting Information). The amount of TKKL formed by module 6-TE is similar to the combined amount of TKL and TKKL formed by module 6-TE when NADPH was added (see Figure S10 in the Supporting Information). This result indicates that the ketoreduction step does not dictate the overall rate of triketide lactone formation. Therefore, we conclude that the rate enhancement observed in module 6–3ACP-TE is primarily due to the engineered tandem ACP domain.
Figure 3.
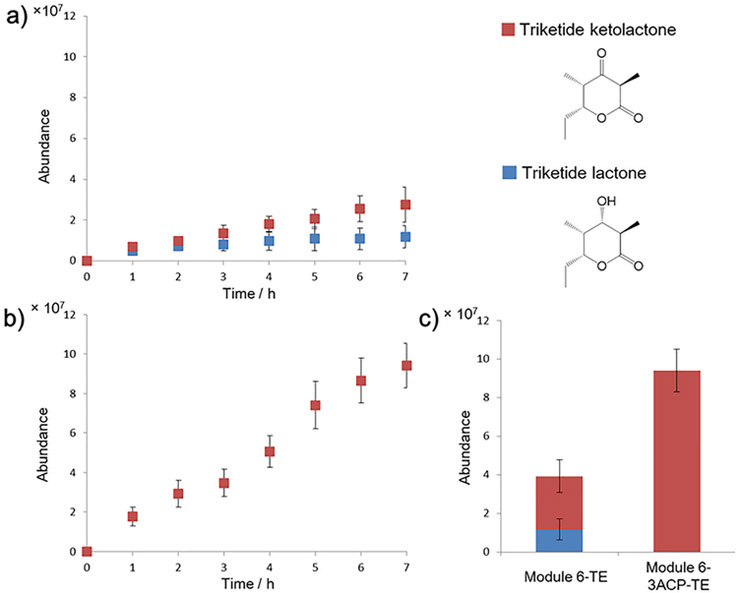
Comparison of the polyketide biosynthesis rate of module 6-TE and module 6–3ACP6-TE: (a) triketide lactone and triketide ketolactone formation by module 6-TE, (b) triketide ketolactone formation by module 6–3ACP-TE, and (c) total amount of polyketide product formed by each protein over 7 h.
Small-Angle X-ray Scattering (SAXS) Analysis of Native and Engineered DEBS Module 6-TE.
Size-exclusion chromatography coupled to SAXS was performed using synchrotron radiation to investigate the structure of module 6-TE and module 6–3ACP-TE. Scattering curves and Guinier plots show good sample quality for both proteins (see Figure S11 in the Supporting Information). Kratky plots for module 6-TE and module 6–3ACP-TE (see Figure 4) approach the baseline at high scattering angles, indicating that both proteins are highly structured. Furthermore, the real-space pair distribution function curves suggest that both proteins have an overall ellipsoidal and disk shape, as indicated by the tail region on the right side and the bulge on the left side of the curve. Module 6-TE has a Dmax value of 214 Å, whereas module 6–3ACP-TE has a Dmax value of 254 Å, indicating that the insertion of two extra ACPs has increased the longest axis dimension of the protein by approximately the combined size of two ACPs.
Figure 4.
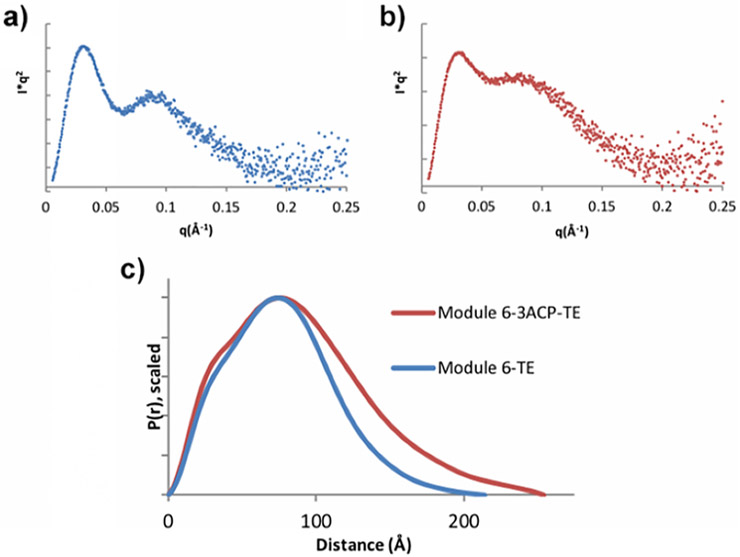
Small-angle X-ray scattering (SAXS) analysis of DEBS module 6-TE and module 6–3ACP-TE: (a) Kratky plot of module 6-TE, (b) Kratky plot of module 6–3ACP-TE, and (c) p(r) of both module 6-TE and module 6–3ACP-TE.
Detailed structural information on full-length PKSs is very limited. Only the SAXS-derived model of DEBS module 3-TE,24 DEBS3,24 DEBS module 2,25 and the cryo-EM derived model of PikAIII26,27 are currently available. According to these studies, DEBS modules have an overall X-shape, which is reminiscent of the porcine FAS architecture,28,29 and they contain two separate reaction chambers. In contrast, PikAIII model is shaped like a horseshoe and contains a single central reaction chamber. Another key difference between these PKS models is the positioning of the ACP domain. In the DEBS module 3-TE and DEBS module 2 models, the ACP domain is positioned at the periphery of the protein. In this configuration, adding two extra ACPs will elongate the length of the protein by the size of two ACPs, which is consistent with our own SAXS data (see Figure 4). In the PikAIII model, ACP is located inside the central reaction chamber when it is not loaded with a substrate. Adding two extra ACPs to this configuration will not change the dimension of the protein, since the additional ACPs will likely remain inside the central chamber. Therefore, we predict module 6–3ACP-TE to have an FAS-like X-shape architecture. However, this model does not explain why KR activity is absent in module 6–3ACP-TE. A complete mechanistic understanding of module 6–3ACP-TE will require high-resolution structural information.
Recently, Hayashi and colleagues showed that inserting two additional ACPs in a PUFA synthase module that natively harbors a 9×ACP domain results in an 2-fold increase in fatty acid production.30 Our study has demonstrated that inserting two extra ACPs to a prototypical type I PKS that originally contains a single ACP results in an ~2.5-fold increase in polyketide production. However, module 6–3ACP-TE does not show any ketoreductase activity, and the reason for this is not clear. A complete understanding of the molecular basis of this defect will require high-resolution structural information on module 6–3ACP-TE which is not available. Interestingly, the KR domain in deep-sea PUFA synthases is positioned downstream of the ACP, whereas the KR domain in module 6–3ACP6-TE is located upstream of the ACP. This difference in domain organization could be why KR activity is lost in 6–3ACP6-TE. Therefore, it will be worthwhile to investigate whether changing the location of the tandem ACP domain, to upstream of KR, will reinstate the original KR activity. Alternatively, installing a longer linker between the individual ACPs may also bring back KR activity, which would provide greater translational and rotational freedom for the constituent ACPs. The full potential of tandem ACP engineering is unknown. PUFA synthase from Schizochytrium sp. contains a total of nine ACPs,13 whereas our 6–3ACP6-TE protein contained just three ACPs. It is conceivable that creating an artificial tandem ACP containing more than three ACPs will lead to further increased product turnover.
METHODS
Chemicals, Plasmids, and Strains.
All reagents were purchased from Sigma–Aldrich (St. Louis, MO), unless stated otherwise. DEBS3-pET21 plasmid, SFP-pET21 plasmid, NDK-SNAc, and E. coli BAP1 strain were gifts from Dr. Chaitan Khosla (Stanford University).
Gene Cloning.
The module 6-te, ks-at-kr, acp6, and te genes were amplified byPCR from DEBS3-pET21 plasmid with Phusion Hot Start DNA polymerase (Thermo, USA). 3×acp6 was custom-synthesized by GenScript (Piscataway, NJ) with codon optimization and then subcloned into pET28 vector with NdeI-EcoRI restriction sites. Module 6-TE-pET28 was constructed by inserting module 6-te into pET-28. Module 6–3ACP-TE-pET28 was constructed by sequentially inserting ks-at-kr, 3×acp6, and te into pET-28.
Gene Expression and Protein Production and Purification.
All proteins were produced in E. coli BAP1 strain. Cell stock was inoculated into 10 mL of LB broth containing 50 μg/mL kanamycin, incubated at 37 °C overnight at 225 rpm. This seed culture was used to inoculate 1 L of LB broth containing 50 μg/mL kanamycin and then incubated at 37 °C for 2–3 h, at 225 rpm. When the OD600 reached 0.6, the culture was cooled on ice for 15–20 min. 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce protein expression and incubated at 18 °C to allow the protein expression. Cells were harvested by centrifugation at 7000 rpm at 4 °C. Harvested cells were transferred to a 50 mL Falcon tube kept at −80 °C until needed. A pellet from 1 L of cell culture was thawed at room temperature (RT) and resuspended in 10 mL of cell lysis buffer (50 mM NaH2PO4: NaOH, pH 7.8, 300 mM NaCl, 10 mM PMSF). Cells were lysed by sonication for 15 min (500 W, 30% amplitude, pulse = 5 s on and 10 s off). Cell debris and other insoluble material were removed by centrifugation for 45 min at 13 000 rpm at 4 °C. The soluble fraction was filtered using a 0.22 μm filter, mixed with 1 mL of Ni-NTA resin (Thermo Scientific, USA), and incubated at 4 °C for 1 h under constant mixing. The resulting resin was washed with wash buffer (50 mM sodium phosphate, pH 7.8, 0.3 M NaCl, 20 mM imidazole), and the target protein was eluted using the elution buffer (50 mM sodium phosphate, pH 7.8, 0.3 M NaCl, 200 mM imidazole).
In Vitro Conversion of ACP from apo- to holo- Form by Recombinant SFP Protein.
A reaction mixture containing 1 mM SFP, 20 mM module 6-TE or module 6–3ACP-TE, 30 mM coenzyme A or 90 mM coenzyme A, respectively, 50 mM Tris-HCl, pH 8.5, 0.3 M NaCl, 10 mM MgCl2, and 5 mM DTT was incubated at 37 °C for 1 h.
In Vitro Production of Triketide Lactone and Ketolactone by Module 6-TE and Module 6–3ACP-TE.
This experiment was performed as described elsewhere with minor modifications.31 The 200 μL reaction mixture contained 5 μM module 6-TE or module 6–3ACP-TE, 1 mM NDK-SNAc, 2.5 mM TCEP, 1 mM EDTA, 0.2 mM methylmalonyl CoA and 4 mM NADPH, 400 mM phosphate adjusted to pH 7.2, and 20% glycerol. The reaction mixture was incubated at RT for different time periods.
LC-MS/ESI Analysis of Triketide Lactone and Ketolactone Production.
LC analysis was performed on a Poroshell 300SB-C18 column (5 μm, 300 Å, 21 mm × 75 mm, Agilent, USA), and eluted with a gradient of CH3CN from 2% to 95% in 0.1% formic acid-H2O in 18 min at a flow rate of 0.2 mL/min. β-Ketoacyl-SNAc (1 mM) was added as the internal standard before every sample injection to normalize variations in chromatography, ionization, and detection. MS/ESI was performed on an Agilent 6550 iFunnel Q-TOF LC/MS System, and the data were analyzed using MassHunter software.
Small-Angle X-ray Scattering (SAXS) Analysis of Module 6-TE and Module 6/3×ACP6-TE.
SAXS data was collected on the Bio-SAXS beamline BL4-2 at Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Laboratory, Menlo Park, CA), using a Pilatus × 1 M detector with a sample-to-detector distance of 2.5 m and a beam energy of 12.4 keV (wavelength, λ = 1.00 Å).32 A 1.5 mm quartz capillary cell that was maintained at 20 °C was used to keep the sample aliquot in the X-ray beam. The momentum transfer (scattering vector) q was defined as q = 4π sin(θ)/λ, where 2θ is the scattering angle. The q scale was calibrated by silver behenate powder diffraction.28 All data were collected by the data acquisition program Blu-ICE using SEC-SAXS mode (see below for more details).31,33 For each image, an integrated transmission beam intensity value was recorded by a photodiode mounted inside the beamstop. Online size-exclusion chromatography solution X-ray scattering (SEC-SAXS) was performed using Thermo Fisher Scientific UltiMate 3000 system with two Superose 6 Increase 3.2/300 columns. The Blu-ICE SEC-SAXS mode was employed for high-throughput tandem SEC-SAXS data collection and subsequent initial analyses; 500 images were recorded with 1 s of exposure every 5 s at a flow rate of 0.04 mL/min. The data processing program SasTool (http://ssrl.slac.stanford.edu/~saxs/analysis/sastool.htm) was used for scaling, using the transmission intensity, azimuthal integration, averaging of individual scattering images, and background subtraction. The first 100 images at the early part of the void volume were averaged and it is assigned as a buffer-scattering profile for the background. After taking the background images, the X-ray shutter was closed until the elution peak of interest in order to keep the sample cell clean from radiation-damaged sample. The automatic SEC-SAXS data processing and analysis pipeline “SECPipe”, which includes real-time consecutive Guinier analysis implemented in the program AUTORG,34 was used for data quality assessment and averaging scattering profiles every five images.
Supplementary Material
ACKNOWLEDGMENTS
DEBS3-pET21 plasmid, SFP-pET21 plasmid, NDK-SNAc, and E. coli BAP1 strain were gifts from Dr. Chaitan Khosla, Stanford University. Use of the Stanford Synchrotron Radiation Light-source, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences (under Contract No. DE-AC02-76SF00515). The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including No. P41GM103393). Research reported in this paper was supported by the National Institute of General Medical Sciences of the National Institutes of Health under linked Award Nos. RL5GM118969, TL4GM118971, and UL1GM118970. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/acschem-bio.8b00896.
Detailed experimental procedure (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Staunton J, and Weissman KJ (2001) Polyketide biosynthesis: a millennium review. Nat. Prod. Rep 18, 380–416. [DOI] [PubMed] [Google Scholar]
- (2).Khosla C, Tang Y, Chen AY, Schnarr NA, and Cane DE (2007) Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem 76, 195–221. [DOI] [PubMed] [Google Scholar]
- (3).Kim CY, Alekseyev VY, Chen AY, Tang Y, Cane DE, and Khosla C (2004) Reconstituting modular activity from separated domains of 6-deoxyerythronolide B synthase. Biochemistry 43, 13892–13898. [DOI] [PubMed] [Google Scholar]
- (4).Harper AD, Bailey CB, Edwards AD, Detelich JF, and Keatinge-Clay AT (2012) Preparative, in vitro biocatalysis of triketide lactone chiral building blocks. ChemBioChem 13, 2200–2203. [DOI] [PubMed] [Google Scholar]
- (5).Rock C, and Jackowski S (1982) Regulation of phospholipid synthesis in Escherichia coli. Composition of the acyl-acyl carrier protein pool in vivo. J. Biol. Chem 257, 10759–10765. [PubMed] [Google Scholar]
- (6).Xu GY, Tam A, Lin L, Hixon J, Fritz CC, and Powers R (2001) Solution structure of B. subtilis acyl carrier protein. Structure 9, 277–287. [DOI] [PubMed] [Google Scholar]
- (7).Findlow SC, Winsor C, Simpson TJ, Crosby J, and Crump MP (2003) Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus. Biochemistry 42, 8423–8433. [DOI] [PubMed] [Google Scholar]
- (8).Zornetzer GA, Fox BG, and Markley JL (2006) Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry 45, 5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Alekseyev VY, Liu CW, Cane DE, Puglisi JD, and Khosla C (2007) Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci 16, 2093–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Perez DR, Leibundgut M, and Wider G (2015) Interactions of the acyl chain with the Saccharomyces cerevisiae acyl carrier protein. Biochemistry 54, 2205–2213. [DOI] [PubMed] [Google Scholar]
- (11).Okuyama H, Orikasa Y, Nishida T, Watanabe K, and Morita N(2007) Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol 73, 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Morita N, Tanaka M, and Okuyama H (2000) Biosynthesis of fatty acids in the docosahexaenoic acid-producing bacterium Moritella marina strain MP-1. Biochem. Soc. Trans 28, 943–945. [PubMed] [Google Scholar]
- (13).Lippmeier JC, Crawford KS, Owen CB, Rivas AA, Metz JG, and Apt KE (2009) Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids 44, 621–630. [DOI] [PubMed] [Google Scholar]
- (14).Jiang H, Zirkle R, Metz JG, Braun L, Richter L, Van Lanen SG, and Shen B (2008) The role of tandem acyl carrier protein domains in polyunsaturated fatty acid biosynthesis. J. Am. Chem. Soc 130, 6336–6337. [DOI] [PubMed] [Google Scholar]
- (15).Trujillo U, Vazquez-Rosa E, Oyola-Robles D, Stagg LJ, Vassallo DA, Vega IE, Arold ST, and Baerga-Ortiz A (2013) Solution structure of the tandem acyl carrier protein domains from a polyunsaturated fatty acid synthase reveals beads-on-a-string configuration. PLoS One 8, e57859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wu N, Kudo F, Cane DE, and Khosla C (2000) Analysis of the Molecular Recognition Features of Individual Modules Derived from the Erythromycin Polyketide Synthase. J. Am. Chem. Soc 122, 4847–4852. [Google Scholar]
- (17).Wu N, Tsuji SY, Cane DE, and Khosla C (2001)Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J.Am. Chem. Soc 123, 6465–6474. [DOI] [PubMed] [Google Scholar]
- (18).Chen AY, Schnarr NA, Kim CY, Cane DE, and Khosla C (2006) Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc 128, 3067–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Klaus M, Ostrowski MP, Austerjost J, Robbins T, Lowry B, Cane DE, and Khosla C (2016) Protein Protein Interactions, not Substrate Recognition, Dominates the Turnover of Chimeric Assembly Line Polyketide Synthases. J. Biol. Chem 291, 16404–16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).America AH, and Cordewener JH (2008) Comparative LC-MS: a landscape of peaks and valleys. Proteomics 8, 731–749. [DOI] [PubMed] [Google Scholar]
- (21).Xie F, Liu T, Qian WJ, Petyuk VA, and Smith RD (2011) Liquid chromatography-mass spectrometry-based quantitative proteomics. J. Biol. Chem 286, 25443–25449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Milac TI, Randolph TW, and Wang P (2012) Analyzing LC-MS/MS data by spectral count and ion abundance: two case studies. Stat. Interface 5, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Abdel-Hamid ME (2000) Comparative LC-MS and HPLC analyses of selected antiepileptics and beta-blocking drugs. Farmaco 55, 136–145. [DOI] [PubMed] [Google Scholar]
- (24).Edwards AL, Matsui T, Weiss TM, and Khosla C (2014) Architectures of whole-module and bimodular proteins from the 6-deoxyerythronolide B synthase. J. Mol. Biol 426, 2229–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Li X, Sevillano N, La Greca F, Deis L, Liu YC, Deller MC, Mathews II, Matsui T, Cane DE, Craik CS, and Khosla C (2018) Structure-Function Analysis of the Extended Conformation of a Polyketide Synthase Module. J. Am. Chem. Soc 140, 6518–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan AR, Håkansson K, Sherman DH, Smith JL, and Skiniotis G (2014) Structure of a modular polyketide synthase. Nature 510, 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Whicher JR, Dutta S, Hansen DA, Hale WA, Chemler JA, Dosey AM, Narayan AR, Håkansson K, Sherman DH, Smith JL, and Skiniotis G (2014) Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature 510, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Maier T, Jenni S, and Ban N (2006) Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science 311, 1258–1262. [DOI] [PubMed] [Google Scholar]
- (29).Maier T, Leibundgut M, and Ban N (2008) The crystal structure of a mammalian fatty acid synthase. Science 321, 1315–1322. [DOI] [PubMed] [Google Scholar]
- (30).Hayashi S, Satoh Y, Ujihara T, Takata Y, and Dairi T (2016) Enhanced production of polyunsaturated fatty acids by enzyme engineering of tandem acyl carrier proteins. Sci. Rep 6, 35441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Martel A, Liu P, Weiss TM, Niebuhr M, and Tsuruta H (2012) An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. J. Synchrotron Radiat 19, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Huang TC, Toraya H, Blanton TN, and Wu Y (1993) X-ray powder diffraction analysis of silver behenate, a possible low-angle diffraction standard. J. Appl. Crystallogr 26, 180–184. [Google Scholar]
- (33).McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, and Kuhn P (2002) Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat 9, 401–406. [DOI] [PubMed] [Google Scholar]
- (34).Petoukhov MV, Konarev PV, Kikhney AG, and Svergun I (2007) ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr 40, s223–s228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.