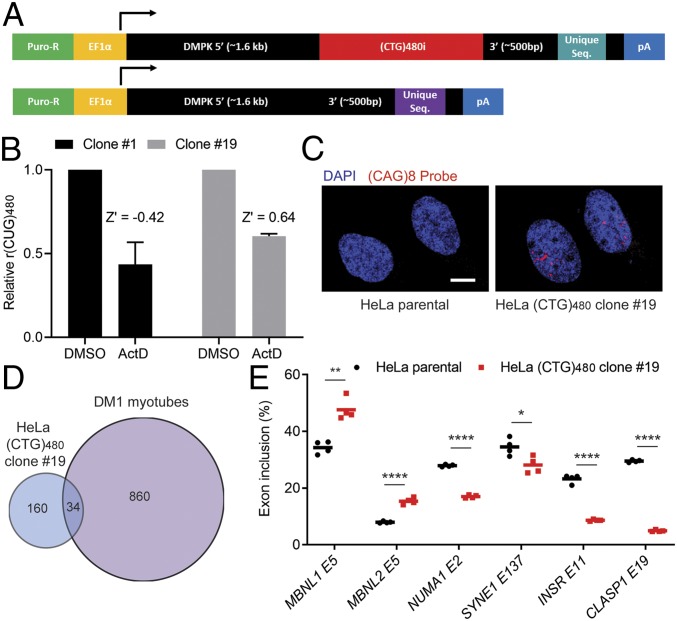
Fig. 1.
Establishing a DM1 HeLa cell line for repeat-selective screening. (A) Schematic of constructs used to generate a stable DM1 HeLa cell model for repeat-selective screening. Constructs are identical in sequence except for the presence of the (CTG)480i, an interrupted repeat tract consisting of 24 modules of [(CTG)20(CTCGA)] and unique sequence (21-bp qPCR probe target and 8-bp barcode). (B) ActD treatment (20 nM) overnight followed by multiplex RT-qPCR [r(CUG)480 relative to r(CUG)0] to determine CTG-selective response in HeLa (CTG)480 cell clones 1 and 19. Data are normalized to DMSO control treatments. Bars represent mean ± SD of 3 biological replicates. Z′ was calculated based on the differential response by C(t) values of (CTG)0 versus (CTG)480 to ActD following the method of Zhang et al. (41). (C) Fluorescence in situ hybridization for rCUGEXP foci using a Cy3-(CAG)8 probe and counterstained with DAPI for nuclei. (Scale bar, 10 µm.) (D) Overlap in alternative cassette exon missplicing events identified by using RNA-seq between HeLa clone 19 and DM1 myoblasts differentiated into myotubes (P < 0.0005, FDR < 0.1, |ΔPSI| > 0.15). (E) RT-PCR isoform analysis of the indicated alternative cassette exon events comparing HeLa (CTG)480 clone 19 to the parental HeLa cell line (mean, n = 4 biological replicates). *P < 0.05; **P < 0.01; ****P < 0.0001.