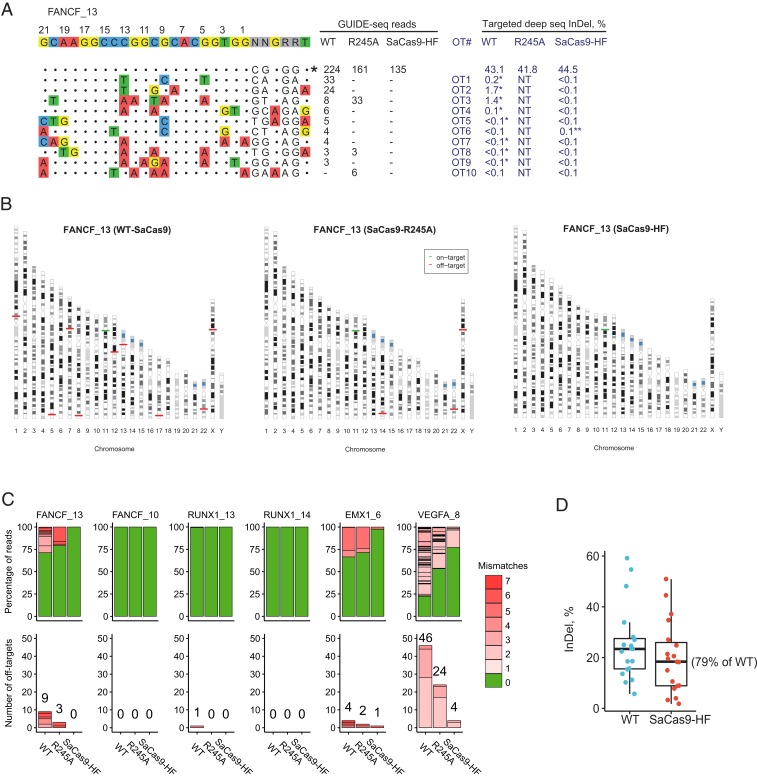
Fig. 4.
Genome-wide editing on- and off-target activity of wild-type SaCas9, R245A, and SaCas9-HF at 6 canonical PAM sites using GUIDE-seq and targeted deep sequencing. (A) On- and off-target activity of the 3 SaCas9s at the target site FANCF_13. On-target site is indicated with “*” at the Right. Mismatched bases in off-target sites with the on-target site are colored. GUIDE-seq read counts for each SaCas9 are listed on the Right. InDel% detected in targeted deep sequencing for the on-target and off-target sites (OT1 through 10) (in dark blue) are listed. “NT” indicates off-target sites not tested using targeted deep sequencing; InDel% marked with “*” indicates edited reads confirmed by IGV visualization; “**” indicates an off-target site prone to false positives (homopolymer G) in targeted deep sequencing and thus its percentage was not calculated. (B) Ideogram presentation of on-target and off-target cleavages detected by GUIDE-seq at FANCF_13. (C) Percentage of edited reads detected by GUIDE-seq at on-target site (green) among total edited reads (Top) and the numbers of off-target sites (ordered by number of mismatches) (Bottom) for each SaCas9. (D) Percentage of InDel reads among all targeted deep sequencing reads at each of the 19 on-target sites, including 6 from C and 13 additional sites (from SI Appendix, Fig. S5), in HEK293T cells caused by WT-SaCas9 or SaCas9-HF editing.