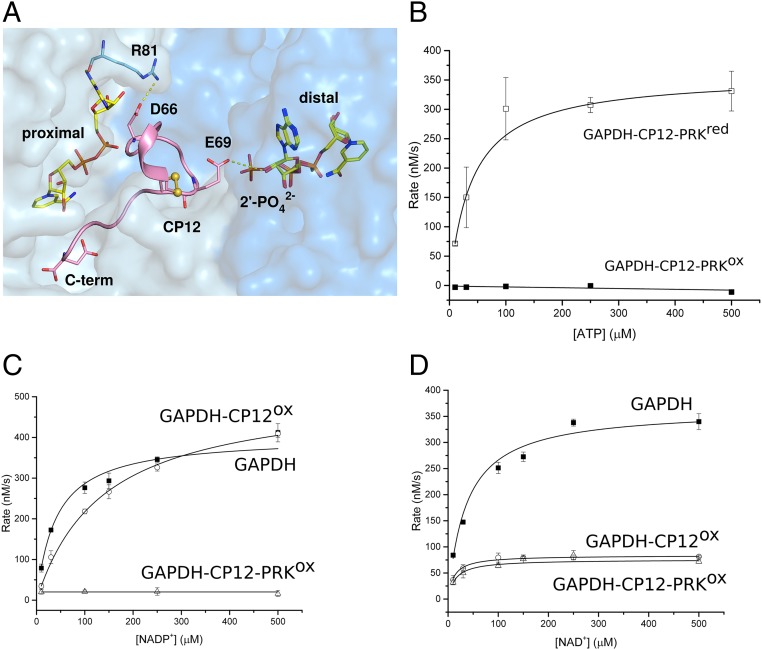
Fig. 1.
Kinetics and coenzyme binding of GAPDH complexes. (A) View of GAPDH (NAD+)-CP12 with adjacent active sites. The CP12-bound (proximal) site makes extensive contacts with both NAD+ and GAPDH. CP12-Glu69 prevents clashes with NADPH binding to the distal site, as it would clash with the 2′-phosphate. (B) PRK activity for the ternary complex treated with either oxidized (GAPDH-CP12-PRKox; ■) or reduced DTT (GAPDH-CP12-PRKred; □). All reactions were measured in triplicate and fitted using Michaelis-Menten kinetics. Rate of NADP+ reduction (C) and NAD+ reduction (D) were measured for 275 µM GAPDH (■), 275 µM GAPDH-CP12 (○), and 63 µM GAPDH-CP12-PRK (△) complexes at increasing concentrations of nucleotide substrate. The maximal velocity (Vmax) with NADP+ is not inhibited in GAPDH-CP12, but is fully inhibited in GAPDH-CP12-PRK. In contrast, the Vmax with NAD+ is equally inhibited by GAPDH-CP12 and GAPDH-CP12-PRK.