FIGURE 4.
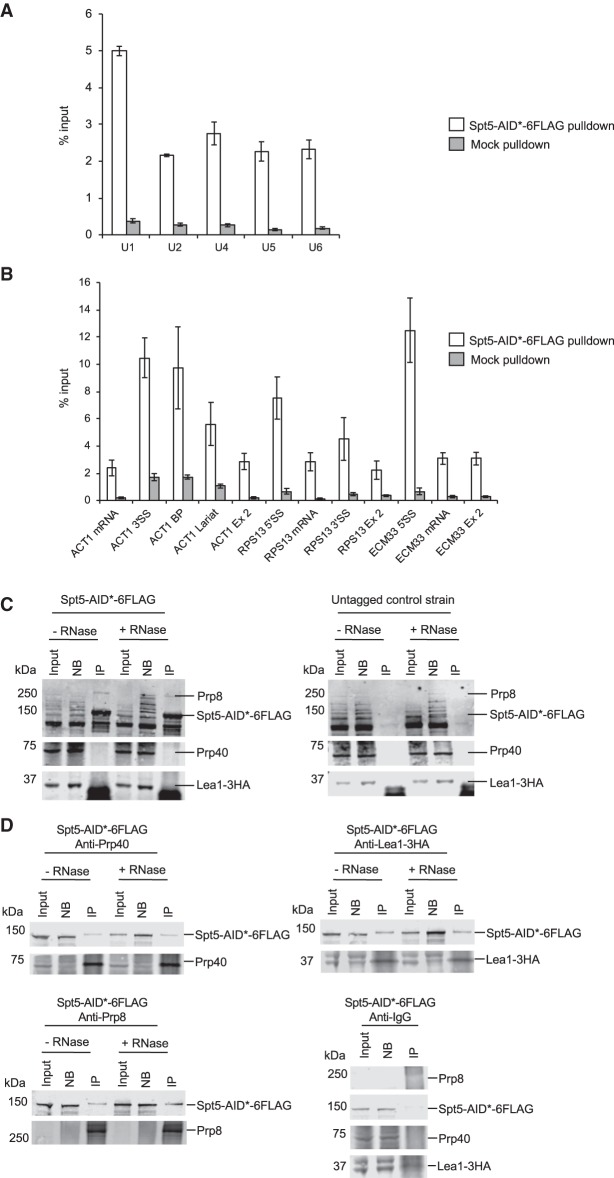
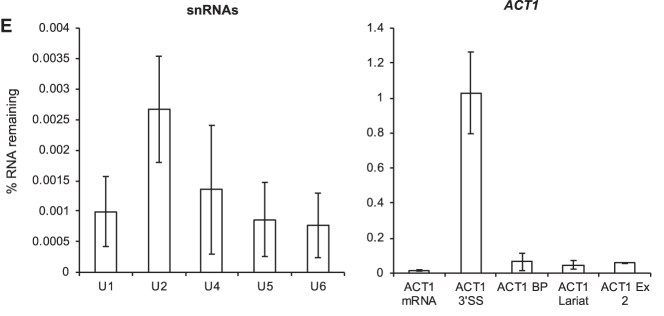
Spt5 interacts with snRNPs. (A) RIP experiment in which Spt5-AID*-6Flag was pulled down followed by RT-qPCR using primers against snRNAs U1, U2, U4, U5, and U6 (white bars). A mock pulldown was also performed (gray bars). Data are normalized to the input. Error bars, standard deviation of three biological replicates. (B) RIP experiment in which Spt5-AID*-6Flag was pulled down followed by RT-qPCR using primers for the intron-containing genes ACT1, RPS13, and ECM33 (white bars). A mock pulldown was also performed (gray bars). Data are normalized as percentage of input (% input). Error bars, standard deviation of three biological replicates. (C) Western blots from a coimmunoprecipitation experiment in which Spt5-AID*-6Flag was pulled down using anti-Flag antibody with or without RNase treatment, blotted and probed with anti-Prp40 (U1 snRNP), anti-Lea-3HA (U2 snRNP), and anti-Prp8 (U5 snRNP) antibodies, then probed with the anti-Flag antibody. Additionally, an untagged Spt5 control strain with Lea1-3HA tagged was used for a pulldown with the Flag antibody as a negative control. Input (10%), nonbound (NB), and immunoprecipitation (IP) samples were loaded. (D) Western blots from coimmunoprecipitation experiments in which the U1 snRNP, U2 snRNP, or U5 snRNP were pulled down using anti-Prp40, anti-HA (for Lea1-HA), or anti-Prp8, respectively, in the Spt5-AID*-6Flag strain with Lea1-3HA tagged. Additionally, a negative rabbit IgG control was included in which rabbit IgG was used for a pulldown with the Spt5-AID*-6Flag strain with Lea1-3HA tagged. Input (10%), nonbound, and immunoprecipitation (IP) samples were loaded. The blot was probed with antibodies against Prp8 (U5 snRNP), Flag (Spt5-AID*-6Flag), Prp40 (U1 snRNP), and HA (Lea1-3HA) (U2 snRNP). (E) RT-qPCR analysis to determine efficiency of RNase treatment of samples used for coimmunoprecipitation in Figure 4C,D. Primers were used against snRNAs U1, U2, U4, U5, and U6 and ACT1 (mRNA, 3′SS, BP, Lariat, and Exon 2). Data are shown as % RNA remaining relative to conditions without RNase treatment. Mean of two biological replicates. Error bars, standard deviation.