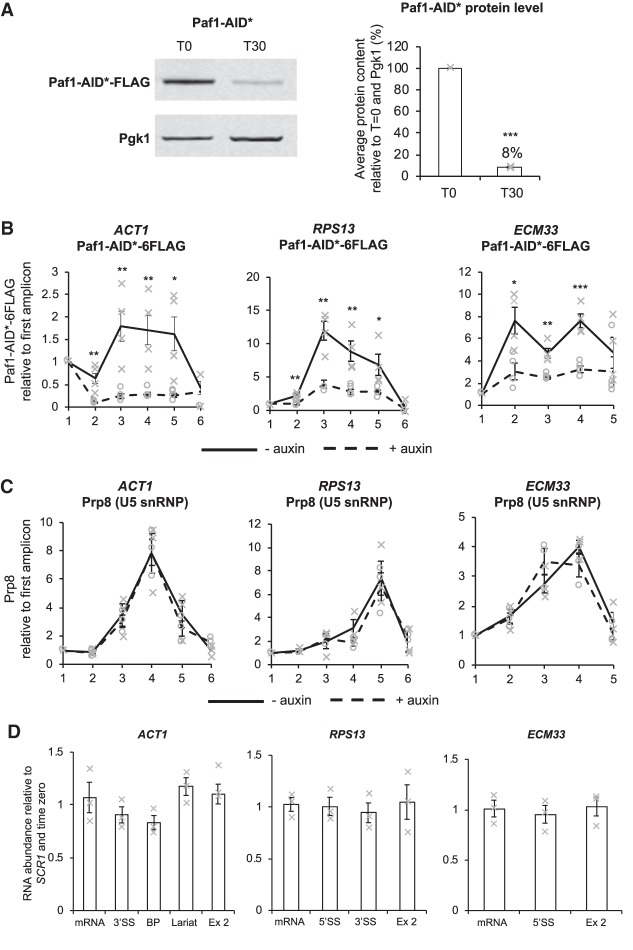
FIGURE 5.
Paf1 depletion does not affect recruitment of U5 snRNP or pre-mRNA splicing. (A) Western blot probed with anti-Flag and anti-Pgk1 as a loading control. T0, samples taken before or T30, 30 min after addition of auxin. Paf1-AID* depletion was quantified and shown as the percentage mean of three biological replicates of T30 relative to T0 values and normalized to the Pgk1 signal. Error bars, standard error of the mean. Gray crosses indicate the individual replicate values. (B) Anti-Flag (Paf1-AID*) and (C) anti-Prp8 (U5 snRNP) ChIP followed by qPCR analysis of the intron-containing genes: ACT1, RPS13, ECM33, 0 min (no auxin; solid black line) and 30 min (+auxin; dashed black line) after auxin addition to depleting Paf1-AID*. X-axes show amplicons used for ChIP-qPCR analysis (see Fig. 1B). Data are presented as mean percentage of input relative to the first amplicon of each gene. Mean of at least three biological replicates. Error bars, standard error of the mean. Asterisks show the statistical significance (Student's unpaired t-test). (*) P < 0.05, (**) P < 0.01, and (***) P < 0.001. Not significant, P > 0.05. Gray crosses indicate the individual replicate values without auxin and β-estradiol and gray circles indicate the individual replicate values 40 min after auxin and β-estradiol addition. (D) RT-qPCR analysis of total RNA from the intron-containing genes ACT1, RPS13, and ECM33 after 30 min of depletion of Paf1-AID*. (A) Normalized to the SCR1 RNAPIII transcript and time zero (no auxin). Primers used detected pre-mRNA (5′SS or BP and 3′SS), lariat (excised intron or intron–exon 2), exon 2 (ex 2), and mRNA (see Fig. 3A for cartoon). Mean of three biological replicates. Error bars, standard error of the mean. Gray crosses indicate the individual replicate values.