FIGURE 1.
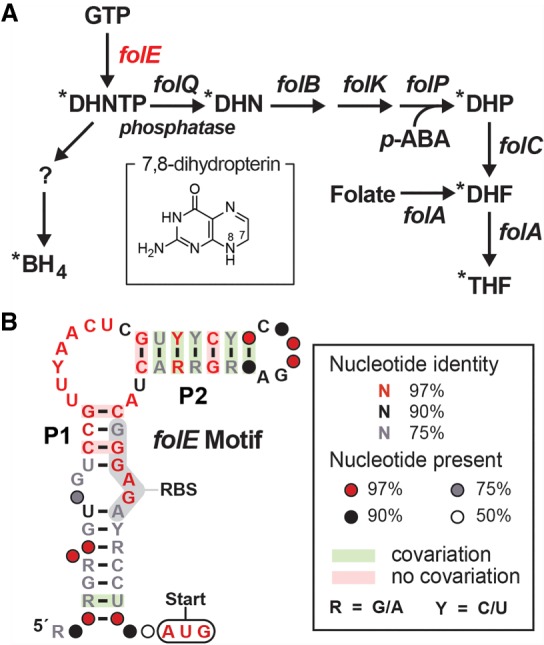
Gene association and consensus model for folE motif RNAs (THF-II riboswitches). (A) Typical tetrahydrofolate (THF) biosynthesis pathway in bacteria, and an additional branch for tetrahydrobiopterin (BH4) biosynthesis. Shared folate and biopterin biosynthesis intermediate: (DHNTP) dihydroneopterin 3′-triphosphate. Exclusive folate biosynthesis intermediates: (DHN) dihydroneopterin, (DHP) dihydroneopterin, (DHF) dihydrofolate. Exclusive BH4 intermediate: “?” represents PTP (6-pyruvoyl-5,6,7,8-tetrahydropterin) or another possible intermediate. All folE motif representatives are associated with the folE gene (red), which codes for the enzyme GTP cyclohydrolase I. Asterisks identify compounds carrying a 7,8-dihydropterin moiety (inset) as produced by the FolE enzyme, or its 5,6,7,8-tetrahydropterin derivative. (B) Updated consensus sequence and secondary structure model for the folE motif, based on 86 unique representatives. The predicted RBS (ribosomal binding site) is highlighted in gray, and the predicted AUG translation start site is circled.
