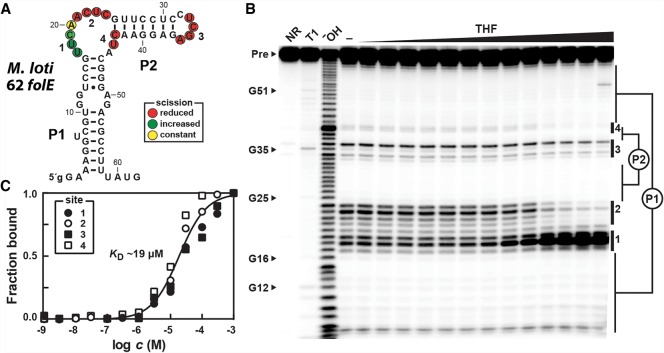
FIGURE 2.
A representative folE motif RNA directly binds THF. (A) Sequence and secondary structure model of the 62 folE RNA construct based on the representative from M. loti. The lowercase letter on the 5′ terminus designates a G residue added to enhance in vitro transcription efficiency. Colored circles identify nucleotides whose phosphodiester linkages undergo scission during in-line probing reactions, wherein THF induces changes as indicated. Data are derived from the autoradiogram in B. (B) PAGE analysis of in-line probing reactions of 5′ 32P-labeled M. loti 62 folE RNA in the absence (−), or the presence of THF ranging in concentration from 1 nM to 1 mM. NR, T1, and OH− indicate no reaction, partial digestion with T1 ribonuclease (cleaves after each G), and partial digestion under alkaline conditions (cleaves after every nucleotide), respectively. Bands corresponding to the precursor RNA (Pre) and several products of RNase T1 digestion (numbered as annotated in A) are indicated. Notable sites of the RNA that undergo modulation upon addition of THF are numbered 1 through 4. (C) Plot of fraction of RNA bound to ligand versus the logarithm of the concentration of THF, as determined based on quantification of band intensity changes at sites 1 through 4 in B. A line representing a theoretical 1:1 binding curve (Hill coefficient of 1) with a KD of 19 µM is overlaid on the data points (R2 value of 0.97) for comparison.