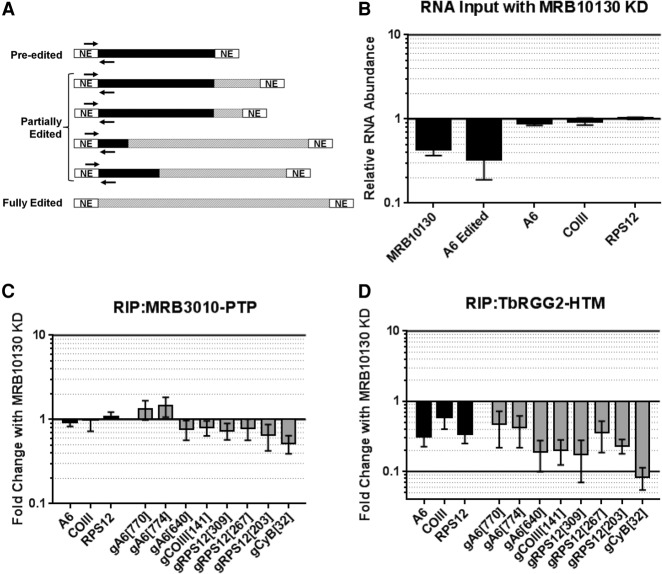
FIGURE 6.
Effect of MRB10130 depletion on RESC protein–RNA interactions. (A) Schematic representation of the qRT-PCR primers used to detect the largest pool of mRNAs for a given transcript in the RNA immunoprecipitations (RIPs). Pre-edited regions are indicated in black, edited RNA is displayed using hashed lines, and never-edited (NE) regions are in white. RIP mRNA primers (indicated with black arrows) hybridize to the 5′ never-edited and 5′-most pre-edited region of the transcripts. RIP primers detect pre-edited and all partially edited RNAs, excluding RNAs that are fully edited. (B) Total cellular RNA from PF cells containing the MRB10130 RNAi construct grown with or without tet for 2 d were analyzed using the RIP qRT-PCR primers. RNA levels were normalized to 18S rRNA levels. The relative RNA abundance represents RNA levels in tet induced cells compared to uninduced. For comparison, primers that detect only fully edited A6 mRNA (as in Fig. 1) are also displayed. qRT-PCR was also used to validate the level of MRB10130 knockdown in the biological replicate experiments (∼40% remaining). (C,D) Comparison of RNA immunoprecipitated with MRB3010-PTP (C) or TbRGG2-HTM (D) grown with or without tet to induce MRB10130 RNAi. Fold change with MRB10130 knockdown represents the RNA levels detected in the RIP from tet-induced cells compared to the RIP from uninduced cells. RNA was detected using RIP mRNA primers described in A and primers designed to detect a subset of gRNAs using qRT-PCR. RNA levels were standardized against 18S rRNA, and numbers represent the mean and standard deviation of two biological replicates with at least eight determinations total.