Abstract
The cell wall is a crucial structural feature in the vast majority of bacteria and comprises a covalently closed network of peptidoglycan (PG) strands. While PG synthesis is important for survival under many conditions, the cell wall is also a dynamic structure, undergoing degradation and remodeling by “autolysins”, enzymes that break down PG. Cell division, for example, requires extensive PG remodeling especially during separation of daughter cells, which depends heavily upon the activity of amidases. However, in V. cholerae, we demonstrate that amidase activity alone is insufficient for daughter cell separation and that lytic transglycosylases RlpA and MltC both contribute to this process. MltC and RlpA both localize to the septum and are functionally redundant under normal laboratory conditions; however, only RlpA can support normal cell separation in low salt media. The division-specific activity of lytic transglycosylases has implications for the local structure of septal PG, suggesting that there may be glycan bridges between daughter cells that cannot be resolved by amidases. We propose that lytic transglycosylases at the septum cleave PG strands that are crosslinked beyond the reach of the highly regulated activity of the amidase and clear PG debris that may block the completion of outer membrane invagination.
Graphical abstract
Vibrio cholerae mutants lacking lytic transglycosylases MltC and RlpA are defective for daughter cell separation. Our results suggest that lytic transglycosylases at the division septum serve as a back-up mechanism to cleave peptidoglycan strands that cannot be cleared by highly-regulated amidase activity and to clear peptidoglycan debris that may block the completion of outer-membrane invagination.
INTRODUCTION
The cell wall is a crucial structural feature for the vast majority of bacteria and is mainly composed of a rigid, yet elastic, covalently-bound network of peptidoglycan (PG) strands. PG has an oligomeric glycan backbone that is assembled by glycosyltransferases (GTs, RodA/FtsW and class A Penicillin Binding Proteins [aPBPs]) (Cho et al., 2016; Leclercq et al., 2017; Zhao et al., 2017; Taguchi et al., 2019) through the polymerization of N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM) heterodimers. These PG strands are crosslinked to adjacent strands primarily by the transpeptidase domains of aPBPs and bPBPs via short peptides attached to the NAM residues, resulting in the strong, mesh-like sacculus. While the rigidity functions to resist bacterial cells’ high internal pressure (Osawa & Erickson, 2018), it restricts processes such as cell growth, division, and insertion of multiprotein trans-envelope complexes such as the flagellum and secretion systems (Nambu et al., 1999; Santin & Cascales, 2017). The cell wall must therefore be a dynamic structure, and indeed undergoes constant remodeling and recycling by PG degradation enzymes collectively known as “autolysins” (T. K. Lee & Huang, 2013).
Autolysins are numerous and diverse, in part owing to the complexity of the substrate on which they act. Many of the different covalent bonds that are found within PG can be cleaved by autolysins in the periplasm, and many of these enzymes are functionally redundant under standard laboratory growth conditions. Functional redundancy has stymied the elucidation of the physiological role of many autolysins, as it makes them inaccessible to many traditional means of assessing gene-phenotype associations, such as analysis of single gene knockouts. The lytic transglycosylases (LTGs), for example, have been exceptionally well-characterized biochemically (Dik et al., 2017), but still relatively little is known about their individual physiological functions. LTGs target PG at the glycosidic bond between NAG and NAM residues and the primary mechanism for this cleavage is a non-hydrolytic, intramolecular cyclization of NAM to form 1,6-anhydroMurNac (anhNAM) (Höltje et al., 1975; Dik et al., 2017; Williams et al., 2018). At least in well-understood model organisms, this signature anhydro “cap” is assumed to be at the end of almost all peptidoglycan strands in vivo (Kraft et al., 1998; Heidrich et al., 2002). Members of the LTG class have been implicated in many cellular processes, including the termination of GT-mediated PG polymerization (Tsui et al., 2016), insertion of secretory apparatuses and flagella (Herlihey & Clarke, 2017; Santin & Cascales, 2017), pathogenesis (Chan et al., 2012), cell envelope integrity (Lamers et al., 2015; Ragland et al., 2017; Crépin et al., 2018), and PG recycling (Cloud & Dillard, 2002).
One process where PG remodeling is particularly important is cell division. Our current understanding of bacterial cell division includes a step in which lateral PG must be remodeled to allow for insertion of a septal wall between daughter cells, followed by cleavage of that septal wall to facilitate daughter cell separation (Potluri et al., 2012; Egan & Vollmer, 2013). Septal PG cleavage by amidases, which cleave off the dipeptide side stem from the NAM residue, is tightly controlled spatiotemporally to ensure that PG degradation is exclusively localized to where it is needed. The amidases are generally assumed to be the main enzymes mediating daughter cell separation, though there is evidence that other autolysins, including LTGs, are pleiotropically involved (Heidrich et al., 2001, 2002; Priyadarshini et al., 2006). E. coli strains lacking LTGs MltABCDE and Slt70, for example, have mild cell separation defects (Heidrich et al., 2002). In addition, Jorgenson, et. al. identified a highly conserved LTG, RlpA, which exhibits septum-specific cleavage activity in Pseudomonas aeruginosa. RlpA is required under low salt conditions, but not during growth in standard laboratory media, suggesting P. aeruginosa encodes at least one redundant septal LTG (Jorgenson et al., 2014). Salmonella enterica similarly appears to require LTGs MltC and MltE for proper daughter cell separation in low salt conditions (Monteiro et al., 2011) and in Neisseria gonorrhea, mutating the LTGs LtgA or LtgC result in daughter cell separation defects (Cloud & Dillard, 2004; Williams et al., 2019). Thus, LTGs appear to play important and often redundant, but poorly-understood roles in septal cleavage in diverse bacteria. In particular, it is unclear whether the separation defects observed in LTG mutants are due to the lack of a septum-specific function of these autolysins, or a general consequence of pleiotropic cell wall damage.
Here we show that in the cholera pathogen, Vibrio cholerae, the two LTGs MltC and RlpA are collectively required for daughter cell separation. Their inactivation results in the generation of cell chains reminiscent of amidase mutants and additional deletion of V. cholerae’s sole amidase exacerbates this chaining defect. Our data suggest that these LTGs fulfill specialized roles in daughter cell separation and have important implications for septal PG architecture in the cholera pathogen.
RESULTS
Simultaneous inactivation of seven LTGs induces a lethal cell separation defect
Autolysins are often functionally redundant. V. cholerae, for example, can tolerate the inactivation of its sole amidase or simultaneous deletion of 5 out of its 6 M23 family endopeptidases (Dörr et al., 2013; Dörr, Davis, et al., 2015), suggesting that redundant autolysins can substitute for each other to sustain at least basic growth processes. In addition to the amidase and the endopeptidases, V. cholerae’s genome encodes 8 predicted LTGs that were identified based on their homology to E. coli LTGs. Whether these enzymes fulfill unique or redundant roles within V. cholerae’s life cycle is unknown. To find new phenotypes associated with LTG deficiency, we endeavored to make sequential deletions in all eight genes. Interestingly, we were able to inactivate six LTGs (mltA, mltB, mltC, mltD, mltF and slt70, leaving only rlpA and mltG intact) with the resulting strain exhibiting only slight morphological aberrations, such as a mild division defect with a corresponding increase in cell length (Fig. 1A, Fig. S1A). Thus, in V. cholerae, RlpA and MltG are cumulatively capable of performing all potentially essential functions of LTGs, at least under laboratory conditions.
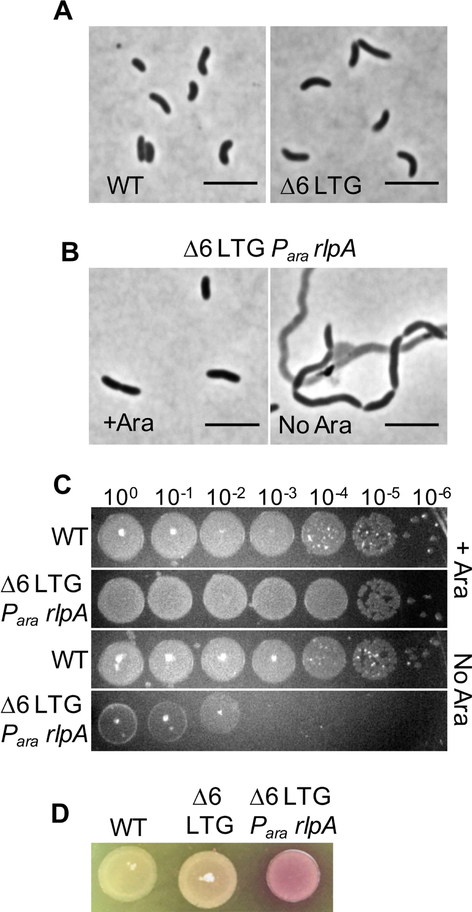
Fig 1. Lytic transglycosylases are collectively required for survival of V. cholerae.
A) WT and Δ6 LTG cultures were grown LB at 37°C and then imaged on an agarose pad. B) RlpA was depleted in the Δ6 LTG background by placing its native promoter under control of arabinose induction and growing in the absence or presence of arabinose (ara). Cells were imaged on an agarose pad and C) spot-plated in 10-fold serial dilutions onto LB+/−0.2% ara, followed by incubation at 30°C for 24hrs. Grid lines = 1cm. D) Lysis was visualized by culturing strains overnight in LB + ara at 37°C, spotting 10μL directly onto an LB+20 μg mL−1 CPRG plate, and incubating 18hrs at 30°C. Scale bars = 5μm. All experiments are representatives of at least two biological replicates.
Though single knockouts of mltG and rlpA could be readily obtained in a wild-type background, (Fig. S1B) we were unable to further delete mltG or rlpA from the Δ6 LTG strain, suggesting that these two represent a minimal set of LTG functions required for viability. To test the phenotypic consequences of loss of RlpA in the Δ6 LTG background, we constructed a strain that placed the native copy of rlpA under control of an arabinose-inducible promoter. Growing this strain in the absence of arabinose, thus depleting RlpA, resulted in the formation of long chains of unseparated cells, many of which had lysed (Fig. 1B). This lysis was also evident in the ~106-fold reduction in plating efficiency after RlpA depletion and dramatic color change when plated on the cell-impermeable β-galactosidase substrate, chlorophenol red-β-D-galactopyranoside (CPRG) (Fig. 1CD), an established readout of cell lysis and inner membrane permeabilization (Paradis-Bleau et al., 2014). The lethal chaining defect seen in the Δ6 LTG strain depleted of RlpA suggests that at least one of the roles of LTGs is an essential function related to septal PG remodeling and/or daughter cell separation that cannot be adequately fulfilled by other autolysins under native conditions. Since a similar depletion strain for MltG did not exhibit a phenotype, we here focused on RlpA for further study; the reason for MltG’s essentiality in the Δ6 background is the subject of future work.
Simultaneous inactivation of RlpA and MltC results in a cell separation defect
Since cell separation defects are not lethal in other V. cholerae autolysin mutants (e.g., amidase mutants [Möll et al., 2014]), we hypothesized that the lethal phenotype of the Δ6 LTG/rlpA depletion strain was in principle separable from the chaining defect. To dissect this further, we assayed different combinations of LTG mutants for cell morphology defects (Fig. S2A). LTGs were generally inactivated by replacing their open reading frame with a scar sequence. However, the gene for RlpA is located within a genomic region containing other important cell wall factors, including rodA and dacA, and so we instead inserted a premature stop codon in position 133 to reduce potential polar effects. Visual inspection of different mutant combinations revealed that a strain carrying both ΔmltC and rlpA::stop (Δ2 LTG) exhibited a pronounced cell separation defect, manifest as long chains in a culture grown to ~OD600 0.8 (Fig 2A). Imaging these chains expressing cytoplasmic GFP from a constitutive promoter revealed clearly separated cytoplasmic spaces. This observation is consistent with photobleaching and fluorescent recovery assays done in E. coli septal autolysin mutants which indicated that cytokinesis was complete in cell chains (Priyadarshini et al., 2007). In contrast to the Δ2 LTG chaining phenotype, the rlpA::stop and ΔmltC single mutants exhibited no morphological differences from the WT in either LB or minimal medium (Fig. S2B). Therefore, MltC and RlpA collectively fulfill a crucial role in daughter cell separation. Despite visible chaining, the Δ2 LTG mutant grew as well as the WT in batch culture (Fig. S2C); however, we did observe a strong, additive motility defect of simultaneous RlpA and MltC inactivation (Fig. S2D). Strong motility defects have been previously associated with other chain-forming mutants, resulting in reduced virulence (Chaput et al., 2016).
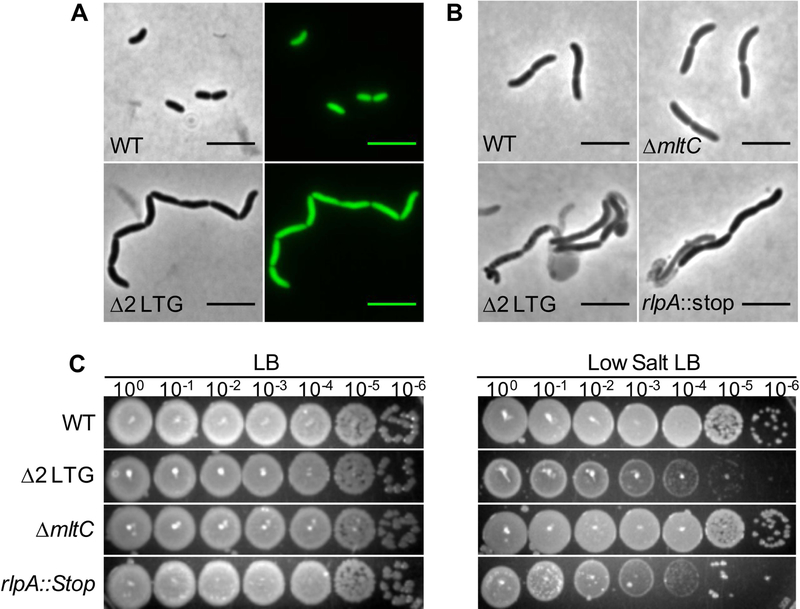
Fig 2. A mutant defective in rlpA and mltC exhibits a chaining defect.
A) WT and Δ2 LTG (ΔmltC rlpA::stop) cultures were grown to OD600 ~0.8 in LB at 37°C and imaged on agarose pads. Cytoplasmic GFP was expressed constitutively from the native lacZ locus. B) The indicated strains were grown for 4 hours in low salt LB (0 mM NaCl) at 37°C and imaged on agarose pads. C) Overnight cultures of the indicated strains grown in LB were diluted in LB and spot-plated in 10-fold serial dilutions on LB and low salt LB and incubated for 18hrs at 30°C.
It has been previously reported that a mutant Pseudomonas aeruginosa lacking RlpA forms unseparated chains of cells when grown in media with low osmolarity (Jorgenson et al., 2014). Similarly, the V. cholerae rlpA::stop single mutant, but not the ΔmltC single mutant, formed chains of cells when grown in low-salt LB medium and exhibited a 1000-fold plating defect on low-salt LB plates (Fig. 2BC). Interestingly, none of these phenotypes were replicated in Escherichia coli. An E. coli ΔrlpA mutant exhibits no obvious morphological defects in LB or low-salt medium (Jorgenson et al., 2014). This was somewhat expected as E. coli RlpA, despite being well-conserved, is predicted to be non-functional as a LTG due to a mutation to a conserved aspartate in the active site as determined by previous alignment with the well-characterized LTG MltA (Fig. S3A) (van Straaten et al., 2005, 2007; Powell et al., 2006; Jorgenson, 2014). Informed by our V. cholerae Δ2 LTG mutant and the S. enterica ΔmltC ΔmltE mutant phenotypes (Monteiro et al., 2011), we made additional, simultaneous mutations in E. coli mltC and mltE, but still observed only a very mild morphological defect (Fig. S3BC). Thus, there are species-specific differences in the utilization of these autolysin homologs.
Δ2 LTG mutations result in outer membrane perturbations
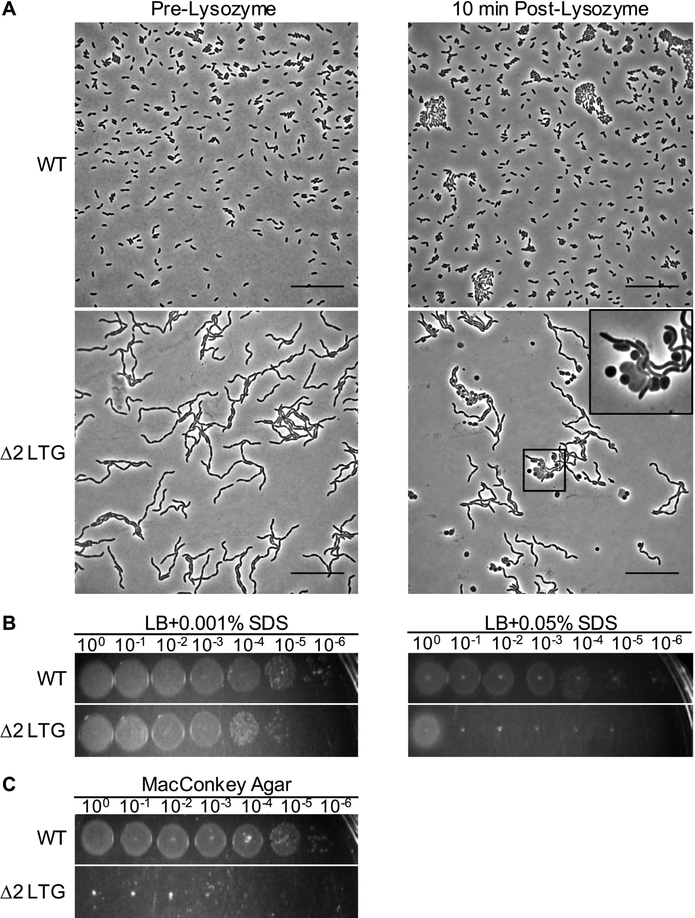
Given that daughter cell separation-defective chains are sensitive to osmotic stress, we also hypothesized that the outer membrane (OM) may be compromised in the Δ2 LTG mutant, as the OM is an important structural and load-bearing component of Gram-negative bacteria (Rojas et al., 2018). Lysozyme typically cannot permeate the OM in sufficient quantities to cause damage to the Gram-negative cell wall. Consistent with this, WT V. cholerae tolerated exposure to 5 mg mL−1 lysozyme without exhibiting signs of morphological defects (Fig. 3A). In contrast, a significant portion of Δ2 LTG mutant cells exhibited a loss of rod shape after just a brief exposure to the same concentration of lysozyme (Fig. 3A). Additionally, Δ2 LTG exhibits a strong plating defect compared to WT on both, LB plates containing sodium dodecyl sulfate (SDS) and MacConkey agar, which contains bile salts (Fig. 3BC). While neither rlpA::stop nor ΔmltC single mutants appeared sensitive to lysozyme or bile salts (Fig. S4AC), rlpA::stop was more sensitive to SDS than the wildtype, though less sensitive than the Δ2 LTG indicating an additive effect of the rlpA and mltC on OM integrity (Fig. S4B). This OM defect suggests that delayed cell separation may present a barrier to proper OM invagination that results in cell envelope perturbations and OM permeabilization.
Fig 3. A mutant defective in rlpA and mltC exhibits an outer membrane defect.
A) Exponential phase cultures of WT and Δ2 LTG were exposed to 5 mg mL−1 lysozyme for 10 min. Scale bars = 25 μm. Insert shows an enhanced field demonstrating partial lysis. B-C) Overnight cultures of WT and Δ2 LTG grown in LB were diluted in LB and spot-plated in 10-fold serial dilutions on LB containing SDS (B) or MacConkey agar and incubated at 18hrs at 30°C (C). All data are representative of at least two biological replicates.
Δ2 LTG and ΔamiB chaining phenotypes are growth phase dependent
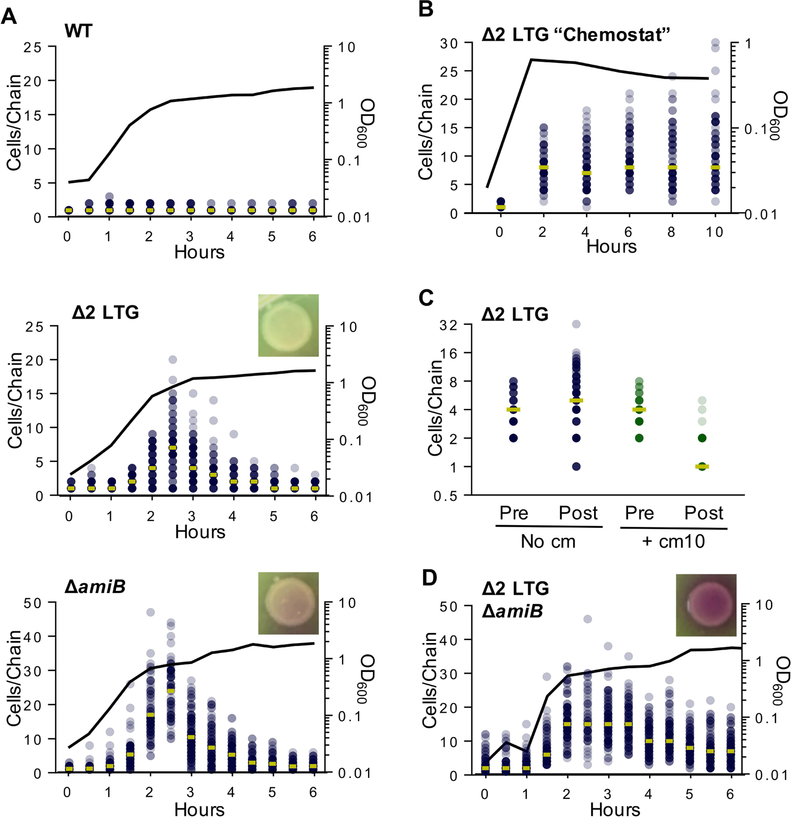
The Δ2 LTG chaining phenotype is reminiscent of a mutant deleted in V. cholerae’s sole amidase, AmiB, which also exhibits chain formation (Möll et al., 2014) due to a well-described daughter cell separation defect (Heidrich et al., 2001, 2002; Priyadarshini et al., 2007; Uehara et al., 2009, 2010). We thus explored other similarities between the two mutants. During our imaging experiments, we noticed that overnight cultures of Δ2 LTG and ΔamiB mutants were devoid of any cell chains, suggesting that chaining was a growth phase-specific phenotype. Single cells of stationary phase Δ2 LTG and ΔamiB were not spontaneous suppressor mutants arising at a high frequency, as redilution into exponential phase always resulted in renewed chain formation and subsequent resolution in stationary phase (Fig. S5). To more precisely define the growth stage that promotes cell chain resolution in these mutants, we imaged the mutant strains versus WT over their growth cycle and quantified the cells per chain at each time point (Fig. 4A). Both Δ2 LTG and ΔamiB exhibited peak chain lengths in mid- to late exponential phase (OD600 ~0.8, median chain length of 8 cells for Δ2 LTG and 24 cells for ΔamiB), followed by a gradual decline in chain length as the cells entered stationary phase.
Fig 4. Chaining defects of septal autolysin mutants depend on fast growth and stochastic resolution.
A) WT, Δ2 LTG (ΔmltC rlpA::stop), and ΔamiB were grown in LB at 37°C and imaged on agarose pads. Cells per chain where manually counted (n >100). Circles represent raw data points of cells/chain (gold bar = median), line graph shows OD600. B) A Δ2 LTG culture was back-diluted every 2 hrs into pre-warmed LB at 37°C to maintain exponential phase and imaged on agarose pads. Analysis was conducted as described for Fig. 4A C) Chloramphenicol (10 μg mL−1, ~10 × MIC) was added to cultures after growth to OD600~0.45 in LB at 37°C and imaged on agarose pads just prior to chloramphenicol addition and 1 hr after addition. Analysis was conducted as described for Fig. 4A. D) Δ2 LTG ΔamiB was grown in LB at 37°C and imaged on agarose pads. Analysis of chain length and CPRG lysis assays were performed as described for Fig. 4A. All data are representative of at least two biological replicates.
The recurring shift between an enrichment of chains in exponential phase and single cells in stationary phase could be explained by stationary phase-specific events, e.g., growth-phase specific lysis cycles or the induction of stationary phase-specific PG remodeling pathways. Alternatively, cell separation could be due to a stochastic process, i.e., reduced septal cleavage activity is initially outpaced by septal PG synthesis during rapid growth, but becomes sufficient for daughter cell separation as division rates slow in stationary phase. Growth phase-dependent chaining has been previously reported for E. coli amidase mutants and it was postulated that single cells were likely generated by lysis of cells within a chain (Heidrich et al., 2001). We tested this hypothesis in V. cholerae by plating autolysin mutants on CPRG. The Δ2 LTG mutant failed to indicate significant CPRG degradation (Fig. 4A), suggesting that lytic elimination of long chains, either by mechanical sheering or cell death, is an unlikely explanation for stationary phase cell separation. The absence of excessive cell debris in microscope images corroborated this observation, as did the WT-equivalent growth rate of this mutant (Fig. S2C). In contrast, the ΔamiB mutant did exhibit a slight color change on CPRG, indicative of higher background lysis, which is consistent with previous observations in E. coli amidase mutants (Heidrich et al., 2001; Priyadarshini et al., 2007). Additionally, this mutant in V. cholerae suffers a slight defect in growth rate when compared to WT (Möll et al., 2014) and Δ2 LTG, suggesting that at least some of the chain resolution of the ΔamiB mutant may depend on lysis (Fig. 4A, Fig. S2C). Thus, while cell lysis may somewhat contribute to apparent chain “resolution” in the amidase mutant, it is unlikely to facilitate the same in the Δ2 LTG mutant.
Δ2 LTG chain resolution is stochastic
We conducted several experiments to help distinguish between chain resolution by stationary phase-induced PG remodeling and stochastic activity of other hydrolases. First, we performed a chemostat-like experiment where Δ2 LTG cells were kept in prolonged exponential phase through periodic back-dilution to maintain an OD600 < 0.6. In a simple scenario, if chain resolution were mediated by a stationary phase-exclusive factor, chains would be expected to elongate ad infinitum when kept in perpetual exponential phase and single cells would no longer be present in the population. However, if other autolysins could stochastically resolve shared septal PG, albeit with a lower efficiency, we would expect an increase in the variation of chain lengths rather than infinite chains. What we observed was more indicative of stochastic resolution. There was some increase in maximum chain length (Fig. 4B), but chains must also have achieved some degree of resolution, as short chains and single cells were still present even after five back-dilutions. These results tentatively suggest that an equilibrium between division and subsequent separation can be achieved in a Δ2 LTG mutant without entering stationary phase.
To test this further, we also surveyed other possible stationary phase-specific factors that could affect cell separation. Culturing Δ2 LTG in the filter-sterilized supernatant of a saturated WT culture failed to promote stationary phase levels of chain resolution (Fig. S6A) as would have been expected should chain resolution be modulated by a secreted compound, for example D-amino acids, which in V. cholerae are exclusively produced in stationary phase (Cava, de Pedro, et al., 2011; Cava, Lam, et al., 2011). We also inactivated the global stationary phase transcriptional regulator, RpoS, in the Δ2 LTG background and found that chain formation during exponential phase and chain resolution during stationary phase were unaffected (Fig. S6B). Finally, we blocked all new protein synthesis in Δ2 LTG grown to exponential phase (OD600~0.4) with chloramphenicol (10μg mL−1, ~15 × minimum inhibitory concentration [MIC]). The majority of chloramphenicol-treated Δ2 LTG chains resolved to single or double cells within an hour of chloramphenicol treatment while untreated chains elongated in this time (Fig. 4C). This indicates that the cells have already translated the necessary enzymes for chain resolution by OD600 0.4 and adds further support that chain resolution in Δ2 LTG can be achieved by reducing the division rate.
These experiments collectively indicate that the enzymes responsible for septal resolution in the absence of MltC and RlpA are expressed in exponential phase and do not depend on stationary phase-specific signals or factors. Therefore, it is likely that redundant housekeeping PG hydrolases or LTGs can mediate daughter cell separation, albeit at a lower efficiency, when the main separation systems are inactivated. We thus hypothesize that chain resolution during transition into stationary phase is the consequence of reduced division rate at this growth stage, which allows other, less efficient, autolysins to “catch up” and separate daughter cells.
Alternative septal resolution factors are insufficient in the cumulative absence of RlpA, MltC, and AmiB
Given the similarities between the Δ2 LTG and ΔamiB mutants and the established dependence of cell separation on the tight spatiotemporal regulation of AmiB (Priyadarshini et al., 2006, 2007; Uehara et al., 2010; Peters et al., 2011; D. C. Yang et al., 2011; Möll et al., 2014), we tested the formal hypothesis that RlpA and MltC directly or indirectly contributed to AmiB recruitment and that the Δ2 LTG chaining defect might thus be due to lack of AmiB localization. A functional AmiB-mCherry fusion localized to septal rings in Δ2 LTG chains, suggesting that this cell separation defect occurs despite proper AmiB localization (Fig. S7AB).
We then investigated the possibly redundant autolytic roles of AmiB, RlpA, and MltC by generating a Δ2 LTG ΔamiB triple mutant and quantifying growth phase-dependent chaining. While this strain was viable, it exhibited a much longer lag phase than the Δ2 LTG or ΔamiB mutants (Fig. S2C) and perhaps more strikingly, chain resolution was incomplete even after 24 hours and gave visual evidence of strong lysis under the microscope as well as on CPRG plates (Fig. 4D, Fig. S5). This increased lysis and failure to completely resolve chains suggest that the amidase and septal LTGs are the principle daughter cell separation systems.
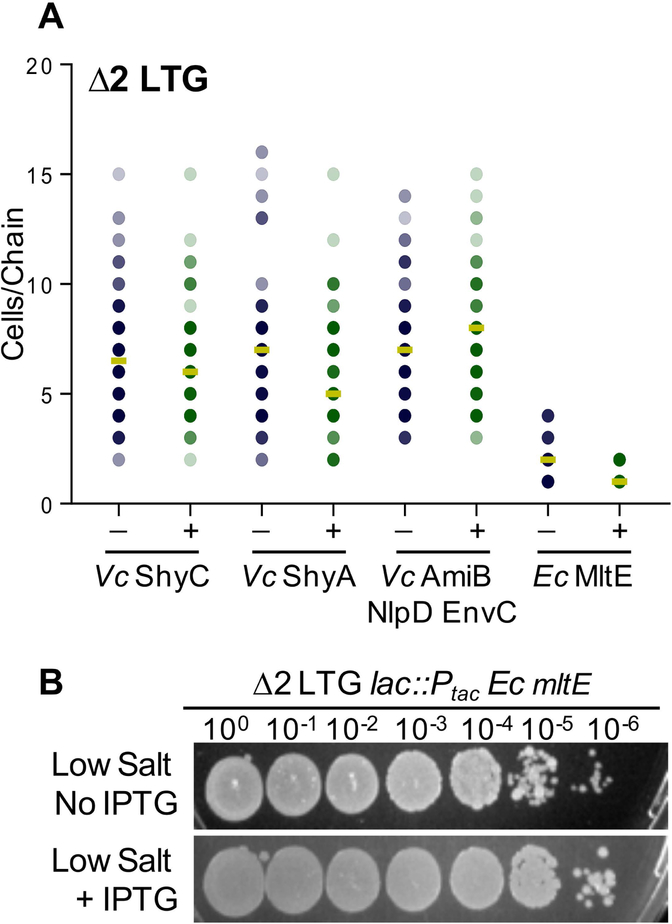
We were interested to learn whether the additive, deleterious effects of the Δ2 LTG and ΔamiB mutations were due to the unique potential functions of LTGs versus amidases at the septum, or if over-expression of AmiB, or other classes autolysins for that matter, could complement the Δ2 LTG chaining defect. Overexpression of V. cholerae’s primary housekeeping endopeptidases, ShyA or ShyC (Dörr et al., 2013), was unable to appreciably reduce chaining in the Δ2 LTG mutant (Fig. 5A) (functional expression of these constructs was validated by their ability to complement a mutant defective in multiple endopeptidases [Dörr et al., 2015]). Similarly, overexpression of V. cholerae’s only amidase, AmiB (co-overexpressed with its activators NlpD and EnvC [Uehara et al., 2009; Yang et al., 2011; Möll et al., 2014]), demonstrated functionality by complementing the ΔamiB mutant (FigS7B), but could not prevent chaining in the Δ2 LTG mutant. Thus, daughter cell separation likely specifically requires LTG glycosidic bond cleavage, rather than general PG hydrolysis by other classes of autolysins.
Fig 5. Lytic transglycosylase activity is required for septal PG resolution.
A) Expression of chromosomal Ptac: shyC, Ptac: shyA, Ptac: amiB nlpD envC, or Ptac: mltEE. coli was induced with 1 mM IPTG in a Δ2 LTG background, grown in LB at 37°C to ~OD600 0.6, and imaged on agarose pads. Analysis was conducted as described for Fig. 4A B) An overnight culture of Δ2 LTG Ptac: mltEE. coli grown in LB was spot-plated in 10-fold serial dilutions on low salt LB +/− 1mM IPTG and incubated for 18hrs at 30°C. All data are representative of at least two biological replicates.
Interestingly, we found that heterologous expression of the E. coli LTG MltE was highly effective at facilitating septal resolution in the Δ2 LTG mutant (Fig. 5A). Addition of 0.2% glucose could not phenotypically suppress MltE complementation of the Δ2 LTG mutant chaining defect (data not shown), suggesting that perhaps leaky expression from Ptac (Rosano & Ceccarelli, 2014) is sufficient for supporting cell separation. MltE has a relatively broad spectrum of PG substrate specificity, shown in vitro to generate products indicative of both endo- and exolytic cleavage on denuded or un-crosslinked muropeptides (M. Lee et al., 2013; Dik et al., 2017; Byun et al., 2018), and has no known homolog in V. cholerae. Overexpression of mltE was not toxic in a WT background, suggesting that it primarily digests septal PG in the V. cholerae Δ2 LTG mutant (Fig. S8). MltE has a strong preference for uncrosslinked PG (M. Lee et al., 2013; Dik et al., 2017; Byun et al., 2018), providing a possible explanation for its lack of general toxicity, since the main body of the cell’s PG is generally crosslinked (Desmarais et al., 2015). Incidentally, this also suggests that septal PG in Δ2 LTG is largely uncrosslinked; which would be consistent with completed or concurrent amidase or endopeptidase activity. MltE was also able to rescue Δ2 LTG growth in low-salt media (Fig. 5B). In combination, these observations suggest that some characteristic of the division septum requires the activity of LTGs over other autolysins to allow for septal resolution and daughter cell separation.
RlpA and MltC are late division proteins
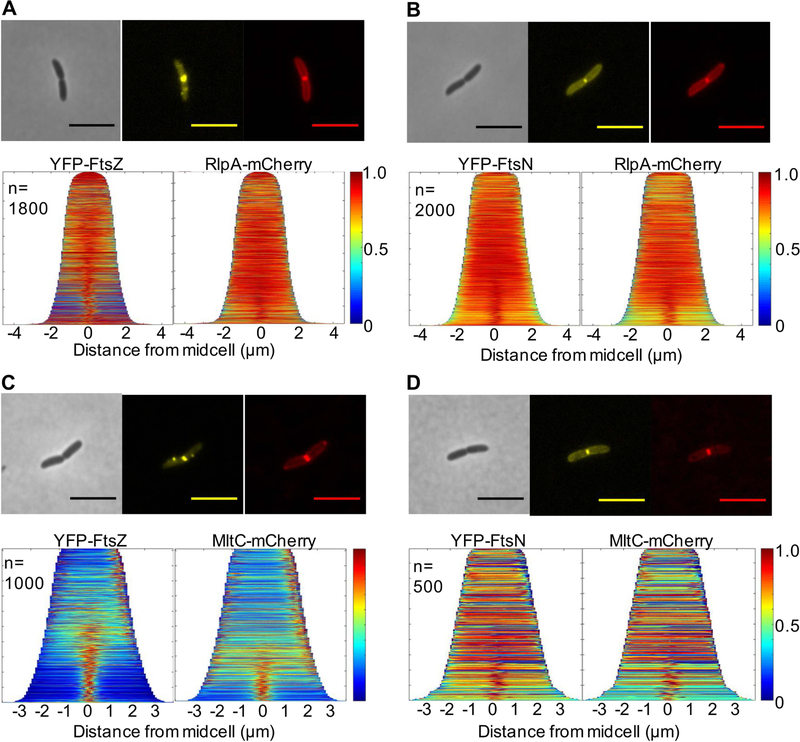
Division proteins are often recruited specifically to midcell. We generated stable, functional (Fig. S9AB) translational C-terminal fusions of MltC and RlpA to mCherry to track localization throughout growth and division. To determine when these LTGs are recruited to the septum, we co-expressed the LTG-mCherry fusions with flourescent fusions of FtsZ, the highly-conserved cytoskeletal element that initiates assembly of the division complex, and FtsN, the late-stage division protein shown to trigger cytokinesis (Egan & Vollmer, 2013). Both RlpA-mCherry and MltC-mCherry clearly localized to the midcell and co-expression of FtsZ-YFP or YFP-FtsN further revealed that both LTGs arrive closer to or after FtsN recruitment to the septum, indicating that RlpA and MltC are likely late division proteins (Fig. 6A–D). RlpA midcell localization is consistent with the published roles of its highly conserved homologues in E. coli and P. aeruginosa (Gerding et al., 2009; Jorgenson et al., 2014; Yahashiri et al., 2015), however, the subcellular localization pattern of MltC homologs in any organism has not been previously reported.
Fig 6. RlpA and MltC are recruited to the septum during late stages of division.
WT cells carrying pBAD33 yfp-ftsN or yfp-ftsZ and pHL100 rlpA-mCherry or mltC-mCherry was grown in M9 + 0.2% glucose supplemented with the appropriate antibiotics at 30°C, induced with 0.2% arabinose and 1mM IPTG after 2hrs, and imaged on agarose pads at OD600~0.15. Demographs of A) RlpA/FtsZ co-localization, B) RlpA/FtsN co-localization, C) MltC/FtsZ co-localization, and D) MltC/FtsN co-localization were generated using Oufti. Scale bars = 5μm. All data are representative of at least two biological replicates.
Outer membrane insertion is not required for MltC and RlpA LTG activity
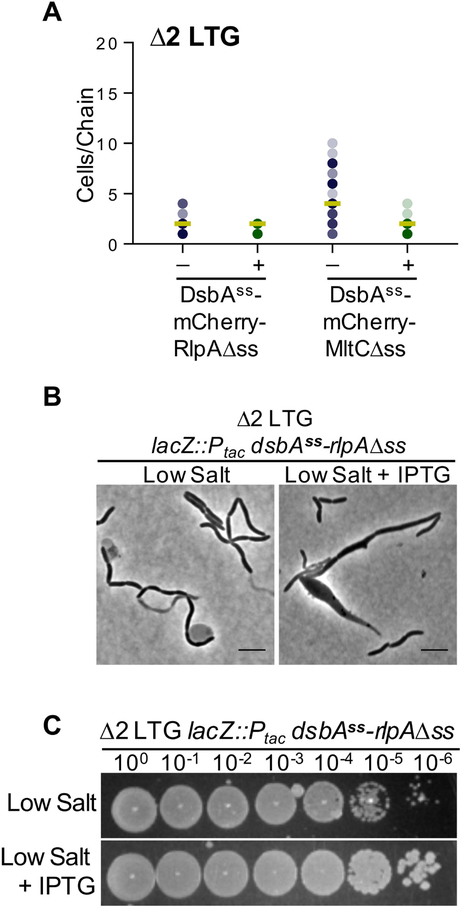
Both LTGs are predicted OM proteins with well-conserved OM target lipoboxes LXGC (Fig. S10A) (Babu et al., 2006), so we postulated that this localization may be important for either their recruitment to the midcell or for their function as septal cleavage enzymes. To test this, we substituted the OM targeting signal sequences of MltC and RlpA N-terminal mCherry fusions with the periplasmic signal sequence of thiol disulfide oxidoreductase, DsbA. Surprisingly, these soluble versions of both proteins were able to complement the Δ2 LTG chaining phenotype in LB (Fig. 7A) yet did so with what appeared to be reduced localization to the midcell (Fig. S10B). Thus, direct OM attachment via lipidation is not essential to the function of RlpA nor MltC in LB, though this does not preclude indirect association via interactions with other OM proteins. However, the functionality of DsbAss-RlpA[18–263] was not absolute under all conditions; expression of this construct in a Δ2 LTG background during growth in low salt LB resulted in the formation of severe morphological aberrations in ~12% of cells (n>400), including short filaments and bulging at the midcell (Fig. 7B). Induction of this defect was not dominant as the DsbAss-RlpA[18–263] mutant did not affect the morphology of WT in low salt LB, nor did over-expression of RlpA-mCherry retaining its native signal sequence affect the morphology of the Δ2 LTG mutant in low salt LB (Fig. S10CD). Despite the apparent division defect induced by soluble DsbAss-RlpA[18–263] expression, the mis-localized protein could still restore growth of the Δ2 LTG mutant on low salt LB (Fig. 7C). The phenotype induced by the DsbAss-RlpA[18–263] mutant in low-salt medium was more similar to filamentous division mutants than to other chaining autolysin mutants. Though we did not explore this further, we thus suspect that RlpA may have conserved roles in division other than septal resolution, roles that may explain why E. coli retains an LTG-deficient homologue of RlpA.
Fig 7. Outer membrane insertion of RlpA and MltC is not required for septal resolution.
A) Expression of chromosomal Ptac: dsbAss-rlpA[18–263] or dsbAss-mltC[42–396] was induced with 1 mM IPTG in a Δ2 LTG background, grown in LB at 37°C to ~OD600 0.6, and imaged on agarose pads. Analysis was conducted as described for Fig. 4A. B) Δ2 LTG Ptac: dsbAss-rlpA[18–263] was grown in low salt LB +/− 1 mM IPTG at 37°C for 4 hrs and imaged on agarose pads. C) Overnight culture of Δ2 LTG Ptac: dsbAss-rlpA[18–263] grown in LB at 37°C was spot-plated in 10-fold serial dilutions onto low salt LB +/− 1mM IPTG and incubated overnight at 30°C. All data are representative of at least two biological replicates.
DISCUSSION
LTG activity plays a conserved role in septal PG resolution
We report here that two LTGs, RlpA and MltC, are septum-specific enzymes with crucial roles in daughter cell separation in V. cholerae. To our knowledge, this is the first report of a physiological role for any housekeeping LTG in the cholera pathogen and the second direct report of a physiological role for MltC (Monteiro et al., 2011). Follow-up studies will be required to determine how septal LTGs interact with the divisome and the PG structural reasons for why MltC and RlpA specifically contribute so significantly to the separation of daughter cells. RlpA and MltC are well-conserved amongst proteobacteria (Artola-Recolons et al., 2014; Yahashiri et al., 2015) yet despite this conservation, mutations in either RlpA (Jorgenson et al., 2014), MltC, or both fail to produce phenotypes in the classic model system E. coli. This study serves to highlight the importance of variety in microbial perspective to understand key processes in bacterial physiology. At the same time, the high conservation of RlpA, especially amongst Gram-negatives (Yunck et al., 2016), and the aberrant morphology of a Δ2 LTG mutant overexpressing dsbAss-rlpA in low salt suggests that septal LTGs may also have other under-appreciated roles in bacterial cell division and daughter cell separation in other Gram-negative pathogens.
A “Cleave and Clear” LTG-dependent model of daughter cell separation
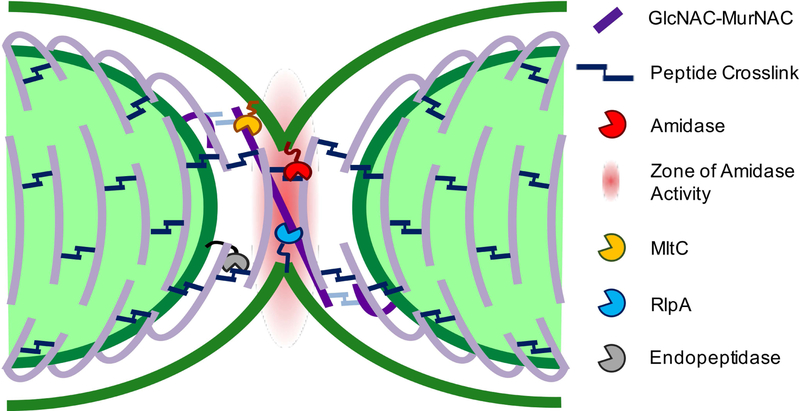
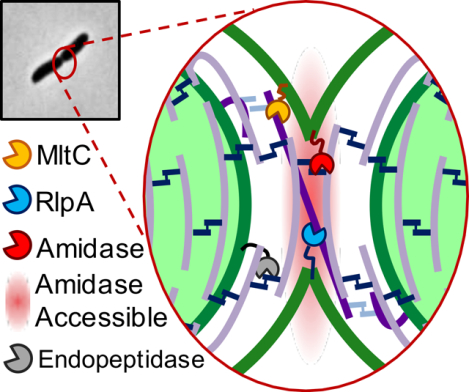
With the addition of RlpA and MltC to the division complex, we can add more detail to the molecular landscape of the septal cell wall. We observed that the chaining defect of the ΔamiB mutant was slightly more severe than the Δ2 LTG mutant, suggesting that the majority of PG shared between daughter cells at the septum is connected by peptide crosslinks that can be removed by amidase activity. However, the chaining phenotype of the Δ2 LTG mutant implies that there is shared septal PG that cannot be resolved by amidase activity. It is ideally expected, from the close association of septal PG synthesis with rotating FtsZ filaments, that PG strands are inserted perpendicular to the long cell axis (Daniel & Errington, 2003; Bisson-Filho et al., 2017; X. Yang et al., 2017). Our findings lead us to speculate that the cell division machinery stochastically generates PG strands that are deposited and/or elongated at a non-ideal angle such that they transect the septal plane to create a bridge between daughter cells (Fig. 8). The PG synthesis machinery may only need to slip from one side of the leading edge of cytokinesis to the other to generate such a strand that is incorporated into both daughter cell sacculi. Since amidase activity is precisely controlled by activators NlpD and EnvC (Möll et al., 2014), the radius of amidase activity is likely highly restricted. Some aberrantly deposited glycan strands might then be crosslinked outside of the range of amidase activity, requiring LTGs as backup enzymes to cleave connecting PG left behind after amidase activity is complete. Consistent with this idea, RlpA in P. aeruginosa acts endolytically on denuded glycan strands (Jorgenson et al., 2014; M. Lee et al., 2017) and has been proposed to follow AmiB to cleave remaining bridging strands. MltC may function to resolve septal PG in two ways. It might also perform a similar function to RlpA, as some endolytic activity has been attributed E. coli MltC (Artola-Recolons et al., 2014). One might also envision that long PG strands between daughter cells, even those that are no longer covalently linked to the PG network, might still present an obstacle to OM invagination. MltC, which in E. coli has been shown to also be processive and exolytic, but inactive on crosslinked PG strands (Artola-Recolons et al., 2014), could be responsible for clearing such debris from the path of OM invagination.
Fig 8. Model of RlpA and MltC requirement for daughter cell separation.
Septal LTGs may remove PG debris that cannot be processed by other autolysins and that impedes the completion of outer membrane invagination.
We suggest that a function to remove aberrant septal PG strands would be particularly important for V. cholerae, which completes the final stages of cell division with astonishing speed (Galli et al., 2017). While the model bacteria E. coli, B. subtilis, and C. crescentus have mature divisomes by 50% of the cell cycle (Aarsman et al., 2005; Gamba et al., 2009; Goley et al., 2011) and E. coli and B. subtilis both encode multiple amidases to assist in daughter cell separation (Heidrich et al., 2001; Firczuk & Bochtler, 2007), V. cholerae encodes a single amidase (Möll et al., 2014) and septation occurs only in the final 10% of its cell cycle (Galli et al., 2017). This is likely to increase the chance for errors to occur, which would create a need to efficiently remove spatial obstructions, including PG aberrantly crosslinked or uncrosslinked PG debris. The absence of a strong chaining phenotype in E. coli ΔmltC ΔrlpA ΔmltE (and the need to delete further LTGs to elicit a weak version of such a phenotype) may indeed reflect this species’ slower division process, resulting in fewer errors. It should also be noted that repression of Ptac is somewhat leaky (Rosano & Ceccarelli, 2014) and that complementation of either the Δ2 LTG or ΔamiB chaining defects occurred readily without induction, which suggests that very little autolytic activity is actually required for separation.
The exponential phase-dependent manner of the ΔamiB and Δ2 LTG chaining phenotype suggests that AmiB and RlpA/MltC functions are redundant with at least one other autolysin whose activity alone is insufficient to sustain daughter cell separation at the fast rate imposed by exponential growth. We hypothesize that RlpA/MltC and AmiB maintain separate functions and that chain resolution in stationary phase is a reflection of each system requiring a distinct autolytic substitute (or group of autolysins) to mediate chain resolution in its absence. For the amidase, this substitute could be D,D or L,D endopeptidases, for LTGs the substitute could be other remaining LTGs. Thus, RlpA, MltC, and AmiB compose the primary group of autolysins responsible for septal resolution and other LTGs or endopeptidases that are secondary to RlpA/MltC or AmiB, respectively, cannot support daughter cell separation in the absence of all three primary septal autolysins. Future work will address the intricate redundancy relationships between these diverse groups of autolysins.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
All V. cholerae strains in this study are derivatives of V. cholerae WT El Tor strain N16961 (Heidelberg et al., 2000) and E. coli strains are derivatives of E. coli K-12 strain MG1655 (Blattner et al., 1997).
Strains were grown at 30°C or 37°C in Luria-Bertani (LB, Fisher BioReagents #BP1425) broth with or without 1.5% NaCl, or in M9 medium containing 0.2% glucose (“M9 Salts,” 2006; “M9 minimal medium (standard),” 2010). When required for selection or plasmid retention, growth media were supplemented with 5-Bromo-4-Chloro-3-Indolyl β-D-Galactopyranoside (X-gal, 40 μg mL−1) streptomycin (200 μg mL−1), kanamycin (5 μg mL−1), carbenicillin (100 μg mL−1), and/or chloramphenicol (5 μg mL−1 for V. cholerae and 20 μg mL−1 for E. coli). Sucrose counter-selection was performed on 10% sucrose LB medium without NaCl. Genes under Ptac or PBAD control were induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) or 0.2% L-arabinose, respectively. For lysis and envelope integrity assays, LB was supplemented with chlorophenol red-β-D-galactopyranoside (CPRG, 20 μg mL−1, Sigma Aldrich # 10884308001), or sodium dodecyl sulfate (SDS), or strains were grown MacConkey Agar (Actero #FCM-133).
At least two replicates were completed per strain and condition for growth curves and growth dependent chaining experiments. For growth curves, strains were inoculated 1:100 into 200 μL of medium from an overnight culture and grown in a Bioscreen growth plate reader (Growth Curves America) at 37°C with random shaking of medium amplitude, and OD600 readings at 5-minute intervals. For chaining experiments with more than three time points, overnight cultures were diluted 1:100 into 200 mL of medium in a 500 mL non-baffled flask and incubated at 37°C with shaking. For experiments with three or fewer time points, overnight cultures were diluted 1:100 into 5 mL of medium in culture tubes.
Construction of Plasmids and Strains
All strains are derivatives of V. cholerae El Tor N16961 or E. coli MG1655. Strains, plasmids, and primers are summarized in Table S1–S4. E. coli DH5α λpir was used for general cloning while E. coli SM10 λpir or MFD λpir (Ferrières et al., 2010) were used for conjugation into V. cholerae. Plasmids were constructed using Gibson assembly (Gibson et al., 2009).
V. cholerae chromosomal in-frame deletions were generated by amplifying 500bp up- and downstream of the gene of interest by PCR, cloning into suicide vector pCVD442 (Donnenberg & Kaper, 1991), and conjugating into V. cholerae N16961 followed by sucrose counter-selection. Briefly, conjugation was performed by mixing and pelleting equal volumes LB overnight culture of plasmid donor E. coli SM10 λpir or MFD λpir strains with recipient V. cholerae, spotting mixed pellet onto an LB (+ 600μM diaminopimelic acid for MFD λpir), and incubating at 37°C for 3 hrs. Single colonies were first selected for on LB + sm200 + vector selection, followed by counter-selection on salt-free LB + 10% sucrose and PCR verification of the deletion.
Ectopic, inducible chromosomal expression of proteins was achieved by cloning the gene(s) of interest open reading frame with either the native 20bp upstream sequence, a strong, consensus ribosome binding site (RBS), or no ribosome binding site into pTD101, a derivative of pJL1 (Miyata et al., 2013) engineered to carry the lac promoter, lacIq, and a multiple cloning site for integration of expression constructs at the native lacZ locus. In particular, pAW51 for polycistronic co-expression amiB, nlpD, and envC, was designed such that amiB was translated from a strong consensus RBS and the 20bp upstream regions containing the native RBS’s of nlpD and envC gene were included between amiB and nlpD or nlpD and envC, respectively, to generate a 3-gene transcriptional fusion under Ptac regulation. pTD101 derivatives were conjugated into V. cholerae as described above for pCVD442.
Depletion of RlpA was accomplished by using suicide vector pAM299 (Möll et al., 2015) to place rlpA under PBAD control at its native locus. pAM299 was conjugated into V. cholerae as a single crossover, without counterselection. For fluorescent localization or detection of proteins by Western blot, genes were cloned into pBAD33 (Guzman et al., 1995) or pHL100 (Dörr, Alvarez, et al., 2015).
E. coli chromosomal in-frame deletions were generated using a combination of λ Red and FLP recombinase systems as previously described (Cherepanov & Wackernagel, 1995; Datsenko & Wanner, 2000; Murphy & Campellone, 2003). Briefly, plasmids pKD3 and pKD4 were used as templates for amplifying cmR and kanR cassettes flanked by FLP recombinase sites with 50bp homology to the up- and downstream regions of the gene of interest. Electrocompetent MG1655 carrying the λ Red recombinase system on pKM208 was transformed with the PCR product amplified from pKD3/4 and recombinants selected for on LB + kan50 or cm20. The gene replacements were moved into a clean MG1655 background by P1 phage transduction (Thomason et al., 2007) and the resulting strain transformed with pCP20 carrying the FLP recombinase. Finally, pCP20 was induced and cured at 37°C and candidates screened for loss of antibiotic resistance and PCR verification of the chromosomal gene deletion.
Microscopy
Cells from liquid culture were imaged without fixation on 0.8% agarose pads containing the same medium from the relevant experiment using a Leica DMi8 inverted microscope. Phase contrast images of chaining mutants were analyzed manually to calculate the number of cells per chain in >100 chains. Raw phase and fluorescent images were analyzed in Oufti (Paintdakhi et al., 2016) using the pre-set E. coli M9 subpixel parameters for calculation of cell width and length and for generation of fluorescent signal localization “demographs.”
Western Blot Analysis
Expression of translational mCherry fusions was induced in WT V. cholerae with 0.2% arabinose for pBAD33 or 1 mM IPTG for pHL100 and lacZ::Ptac in LB and grown to OD600~0.6. Cells were harvested by centrifugation (9500 g, 15 min) at room temperature and resuspended in 1% SDS + 10 mM dithiothreitol (DTT) lysis buffer. Resuspended cells were incubated at 95°C for 3 min, then sonicated 4 × 5 seconds at 20% amplitude. Standard Western blots against mCherry were performed using polyclonal mCherry antibody (Genetex #GTX59788) and detection by IRDye 800CW secondary antibody (Li-cor #926–32211). After imaging for mCherry, the same blots were then re-incubated with monoclonal RpoA antibody (BioLegend # 663104) detected by IRDye 800CW secondary antibody on an Odyssey CLx imaging device (Li-cor).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matthew Jorgenson (University of Arkansas for Medical Sciences) and all members of the Dörr lab for helpful discussion. We also thank the Waldor lab for sharing plasmids pBAD33 yfp-ftsZ and pBAD33 yfp-ftsN as well as the faculty, staff, and students at the Weill Institute for Cell and Molecular Biology (WICMB) for sharing equipment and resources. Research in the Dörr lab is supported by the NIH/NIAID (1R01 AI143704-01).
DATA SHARING
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, & den Blaauwen T (2005). Maturation of the Escherichia coli divisome occurs in two steps. Molecular Microbiology, 55(6), 1631–1645. 10.1111/j.1365-2958.2005.04502.x [DOI] [PubMed] [Google Scholar]
- Artola-Recolons C, Lee M, Bernardo-García N, Blázquez B, Hesek D, Bartual SG, … Hermoso JA (2014). Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase MltC of Escherichia coli. ACS Chemical Biology, 9(9), 2058–2066. 10.1021/cb500439c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, & Sankaran K (2006). A Database of Bacterial Lipoproteins (DOLOP) with Functional Assignments to Predicted Lipoproteins. Journal of Bacteriology, 188(8), 2761 LP – 2773. 10.1128/JB.188.8.2761-2773.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, … Garner EC (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science, 355(6326), 739–743. 10.1126/science.aak9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, … Shao Y (1997). The Complete Genome Sequence of Escherichia coli K-12. Science, 277(5331), 1453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- Byun B, Mahasenan KV, Dik DA, Marous DR, Speri E, Kumarasiri M, … Mobashery S (2018). Mechanism of the Escherichia coli MltE lytic transglycosylase, the cell-wall-penetrating enzyme for Type VI secretion system assembly. Scientific Reports, 8(1), 4110 10.1038/s41598-018-22527-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, & Waldor MK (2011). Distinct pathways for modification of the bacterial cell wall by non‐canonical <span class="sc">d</span>‐amino acids. The EMBO Journal, 30(16), 3442 LP – 3453. 10.1038/emboj.2011.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, Lam H, de Pedro MA, & Waldor MK (2011). Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cellular and Molecular Life Sciences, 68(5), 817–831. 10.1007/s00018-010-0571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Hackett KT, & Dillard JP (2012). The Lytic Transglycosylases of Neisseria gonorrhoeae. Microbial Drug Resistance, 18(3), 271–279. 10.1089/mdr.2012.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaput C, Ecobichon C, Pouradier N, Rousselle J-C, Namane A, & Boneca IG (2016). Role of the N-Acetylmuramoyl-l-Alanyl Amidase, AmiA, of Helicobacter pylori in Peptidoglycan Metabolism, Daughter Cell Separation, and Virulence. Microbial Drug Resistance, 22(6), 477–486. 10.1089/mdr.2016.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, & Wackernagel W (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene, 158(1), 9–14. [DOI] [PubMed] [Google Scholar]
- Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PDA, Suh H, … Bernhardt TG (2016). Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nature Microbiology, 1(10), 16172 Retrieved from 10.1038/nmicrobiol.2016.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, & Dillard JP (2002). A Lytic Transglycosylase of Neisseria gonorrhoeae Is Involved in Peptidoglycan-Derived Cytotoxin Production. Infection and Immunity, 70(6), 2752–2757. 10.1128/IAI.70.6.2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud KA, & Dillard JP (2004). Mutation of a Single Lytic Transglycosylase Causes Aberrant Septation and Inhibits Cell Separation of Neisseria gonorrhoeae. Journal of Bacteriology, 186(22), 7811–7814. 10.1128/JB.186.22.7811-7814.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crépin S, Ottosen EN, Peters K, Smith SN, Himpsl SD, Vollmer W, & Mobley HLT (2018). The lytic transglycosylase MltB connects membrane homeostasis and in vivo fitness of Acinetobacter baumannii. Molecular Microbiology, 109(6), 745–762. 10.1111/mmi.14000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, & Errington J (2003). Control of Cell Morphogenesis in Bacteria: Two Distinct Ways to Make a Rod-Shaped Cell. Cell, 113(6), 767–776. 10.1016/S0092-8674(03)00421-5 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, & Wanner BL (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences, 97(12), 6640 LP – 6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarais SM, Tropini C, Miguel A, Cava F, Monds RD, de Pedro MA, & Huang KC (2015). High-throughput, Highly Sensitive Analyses of Bacterial Morphogenesis Using Ultra Performance Liquid Chromatography. Journal of Biological Chemistry, 290(52), 31090–31100. 10.1074/jbc.M115.661660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik DA, Marous DR, Fisher JF, & Mobashery S (2017). Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Critical Reviews in Biochemistry and Molecular Biology, 52(5), 503–542. 10.1080/10409238.2017.1337705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, & Kaper JB (1991). Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infection and Immunity, 59(12), 4310–4317. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1937792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Alvarez L, Delgado F, Davis BM, Cava F, & Waldor MK (2015). A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta-lactam tolerance. Proceedings of the National Academy of Sciences, 113(2), 404–409. 10.1073/pnas.1520333113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Cava F, Lam H, Davis BM, & Waldor MK (2013). Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae. Molecular Microbiology, 89(5), 949–962. 10.1111/mmi.12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Davis BM, & Waldor MK (2015). Endopeptidase-Mediated Beta Lactam Tolerance. PLOS Pathogens, 11(4), 1–16. 10.1371/journal.ppat.1004850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, & Vollmer W (2013). The physiology of bacterial cell division. Annals of the New York Academy of Sciences, 1277(1), 8–28. 10.1111/j.1749-6632.2012.06818.x [DOI] [PubMed] [Google Scholar]
- Ferrières L, Hémery G, Nham T, Guérout A-M, Mazel D, Beloin C, & Ghigo J-M (2010). Silent Mischief: Bacteriophage Mu Insertions Contaminate Products of Escherichia coli Random Mutagenesis Performed Using Suicidal Transposon Delivery Plasmids Mobilized by Broad-Host-Range RP4 Conjugative Machinery. Journal of Bacteriology, 192(24), 6418–6427. 10.1128/JB.00621-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk M, & Bochtler M (2007). Folds and activities of peptidoglycan amidases. FEMS Microbiology Reviews, 31(6), 676–691. 10.1111/j.1574-6976.2007.00084.x [DOI] [PubMed] [Google Scholar]
- Galli E, Paly E, & Barre F-X (2017). Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last 10% of the cell cycle. Scientific Reports, 7(December 2016), 1–11. 10.1038/srep44505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba P, Veening J-W, Saunders NJ, Hamoen LW, & Daniel RA (2009). Two-Step Assembly Dynamics of the Bacillus subtilis Divisome. Journal of Bacteriology, 191(13), 4186–4194. 10.1128/JB.01758-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, & de Boer PAJ (2009). Self-Enhanced Accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR Domain (DamX, DedD, and RlpA) in Escherichia coli Cell Constriction. Journal of Bacteriology, 191(24), 7383–7401. 10.1128/JB.00811-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, & Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6, 343–345. Retrieved from 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goley ED, Yeh Y-C, Hong S-H, Fero MJ, Abeliuk E, McAdams HH, & Shapiro L (2011). Assembly of the Caulobacter cell division machine. Molecular Microbiology, 80(6), 1680–1698. 10.1111/j.1365-2958.2011.07677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, & Beckwith J (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology, 177(14), 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, … Fraser CM (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature, 406(6795), 477–483. 10.1038/35020000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, … Höltje J-V (2001). Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Molecular Microbiology, 41(1), 167–178. 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H, & Höltje J-V (2002). Effects of Multiple Deletions of Murein Hydrolases on Viability, Septum Cleavage, and Sensitivity to Large Toxic Molecules in Escherichia coli. Journal of Bacteriology, 184(22), 6093–6099. 10.1128/JB.184.22.6093-6099.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlihey FA, & Clarke AJ (2017). Controlling Autolysis During Flagella Insertion in Gram-Negative Bacteria. In M. Z. Atassi (Ed.), Protein Reviews: Volume 17 (pp. 41–56). 10.1007/5584_2016_52 [DOI] [PubMed] [Google Scholar]
- Höltje JV, Mirelman D, Sharon N, & Schwarz U (1975). Novel type of murein transglycosylase in Escherichia coli. Journal of Bacteriology, 124(3), 1067–1076. Retrieved from http://jb.asm.org/content/124/3/1067.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson MA (2014). A tale of two RLPAs : studies of cell division in Escherichia coli and Pseudomonas aeruginosa. Iowa Research Online. Retrieved from https://ir.uiowa.edu/etd/1342. [Google Scholar]
- Jorgenson MA, Chen Y, Yahashiri A, Popham DL, & Weiss DS (2014). The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Molecular Microbiology, 93(1), 113–128. 10.1111/mmi.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft AR, Templin MF, & Höltje J-V (1998). Membrane-Bound Lytic Endotransglycosylase in Escherichia coli. Journal of Bacteriology, 180(13), 3441–3447. Retrieved from http://jb.asm.org/content/180/13/3441.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers RP, Nguyen UT, Nguyen Y, Buensuceso RNC, & Burrows LL (2015). Loss of membrane-bound lytic transglycosylases increases outer membrane permeability and β-lactam sensitivity in Pseudomonas aeruginosa. MicrobiologyOpen, 4(6), 879–895. 10.1002/mbo3.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Derouaux A, Olatunji S, Fraipont C, Egan AJF, Vollmer W, … Terrak M (2017). Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Scientific Reports, 7, 43306 Retrieved from 10.1038/srep43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Dik DA, Fishovitz J, Lastochkin E, Boggess B, … Mobashery S (2017). From Genome to Proteome to Elucidation of Reactions for All Eleven Known Lytic Transglycosylases from Pseudomonas aeruginosa. Angewandte Chemie International Edition, 56(10), 2735–2739. 10.1002/anie.201611279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, & Mobashery S (2013). Reactions of All Escherichia coli Lytic Transglycosylases with Bacterial Cell Wall. Journal of the American Chemical Society, 135(9), 3311–3314. 10.1021/ja309036q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, & Huang KC (2013). The role of hydrolases in bacterial cell-wall growth. Current Opinion in Microbiology, 16(6), 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M9 minimal medium (standard). (2010). Cold Spring Harbor Protocols, 2010(8), pdb.rec12295. 10.1101/pdb.rec12295 [DOI] [Google Scholar]
- M9 Salts. (2006). Cold Spring Harbor Protocols, 2006(1), pdb.rec614. 10.1101/pdb.rec614 [DOI] [Google Scholar]
- Miyata ST, Unterweger D, Rudko SP, & Pukatzki S (2013). Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic Vibrio cholerae. PLOS Pathogens, 9(12), e1003752 Retrieved from 10.1371/journal.ppat.1003752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möll A, Dörr T, Alvarez L, Chao MC, Davis BM, Cava F, & Waldor MK (2014). Cell Separation in Vibrio cholerae Is Mediated by a Single Amidase Whose Action Is Modulated by Two Nonredundant Activators. Journal of Bacteriology, 196(22), 3937–3948. 10.1128/JB.02094-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möll A, Dörr T, Alvarez L, Davis BM, Cava F, & Waldor MK (2015). A D, D-carboxypeptidase is required for Vibrio cholerae halotolerance. Environmental Microbiology, 17(2), 527–540. 10.1111/1462-2920.12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro C, Fang X, Ahmad I, Gomelsky M, & Römling U (2011). Regulation of Biofilm Components in Salmonella enterica Serovar Typhimurium by Lytic Transglycosylases Involved in Cell Wall Turnover. Journal of Bacteriology, 193(23), 6443 LP – 6451. 10.1128/JB.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, & Campellone KG (2003). Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Molecular Biology, 4, 1–12. 10.1186/1471-2199-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu T, Minamino T, Macnab RM, & Kutsukake K (1999). Peptidoglycan-Hydrolyzing Activity of the FlgJ Protein, Essential for Flagellar Rod Formation in Salmonella typhimurium. Journal of Bacteriology, 181(5), 1555–1561. Retrieved from http://jb.asm.org/content/181/5/1555.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, & Erickson HP (2018). Turgor Pressure and Possible Constriction Mechanisms in Bacterial Division. Frontiers in Microbiology, 9, 111 10.3389/fmicb.2018.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, & Jacobs-Wagner C (2016). Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Molecular Microbiology, 99(4), 767–777. 10.1111/mmi.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Kritikos G, Orlova K, Typas A, & Bernhardt TG (2014). A Genome-Wide Screen for Bacterial Envelope Biogenesis Mutants Identifies a Novel Factor Involved in Cell Wall Precursor Metabolism. PLOS Genetics, 10(1), e1004056 Retrieved from 10.1371/journal.pgen.1004056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Dinh T, & Bernhardt TG (2011). A Fail-Safe Mechanism in the Septal Ring Assembly Pathway Generated by the Sequential Recruitment of Cell Separation Amidases and Their Activators. Journal of Bacteriology, 193(18), 4973–4983. 10.1128/JB.00316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potluri L-P, Kannan S, & Young KD (2012). ZipA Is Required for FtsZ-Dependent Preseptal Peptidoglycan Synthesis prior to Invagination during Cell Division. Journal of Bacteriology, 194(19), 5334–5342. 10.1128/JB.00859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AJ, Liu Z-J, Nicholas RA, & Davies C (2006). Crystal Structures of the Lytic Transglycosylase MltA from N.gonorrhoeae and E.coli: Insights into Interdomain Movements and Substrate Binding. Journal of Molecular Biology, 359(1), 122–136. [DOI] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, & Young KD (2007). Role of Peptidoglycan Amidases in the Development and Morphology of the Division Septum in Escherichia coli. Journal of Bacteriology, 189(14), 5334–5347. 10.1128/JB.00415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, Popham DL, & Young KD (2006). Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. Journal of Bacteriology, 188(15), 5345–5355. 10.1128/JB.00476-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland SA, Schaub RE, Hackett KT, Dillard JP, & Criss AK (2017). Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cellular Microbiology, 19(3), e12662 10.1111/cmi.12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, … Huang KC (2018). The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature, 559(7715), 617–621. 10.1038/s41586-018-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, & Ceccarelli EA (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Frontiers in Microbiology, 5, 172 10.3389/fmicb2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin YG, & Cascales E (2017). Domestication of a housekeeping transglycosylase for assembly of a Type VI secretion system. EMBO Reports, 18(1), 138–149. 10.15252/embr.201643206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, … Walker S (2019). FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nature Microbiology, 4(4), 587–594. 10.1038/s41564-018-0345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, & Court DL (2007). E. coli Genome Manipulation by P1 Transduction . Current Protocols in Molecular Biology, 79(1), 1.17.1–1.17.8. 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- Tsui H-CT, Zheng JJ, Magallon AN, Ryan JD, Yunck R, Rued BE, … Winkler ME (2016). Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Molecular Microbiology, 100(6), 1039–1065. 10.1111/mmi.13366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, & Bernhardt TG (2009). LytM-Domain Factors Are Required for Daughter Cell Separation and Rapid Ampicillin-Induced Lysis in Escherichia coli. Journal of Bacteriology, 191(16), 5094–5107. 10.1128/JB.00505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, & Bernhardt TG (2010). Daughter cell separation is controlled by cytokinetic ring‐activated cell wall hydrolysis. The EMBO Journal, 29(8), 1412 LP – 1422. 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten KE, Barends TRM, Dijkstra BW, & Thunnissen A-MWH (2007). Structure of Escherichia coli Lytic Transglycosylase MltA with Bound Chitohexaose: IMPLICATIONS FOR PEPTIDOGLYCAN BINDING AND CLEAVAGE. Journal of Biological Chemistry, 282(29), 21197–21205. 10.1074/jbc.M701818200 [DOI] [PubMed] [Google Scholar]
- van Straaten KE, Dijkstra BW, Vollmer W, & Thunnissen A-MWH (2005). Crystal Structure of MltA from Escherichia coli Reveals a Unique Lytic Transglycosylase Fold. Journal of Molecular Biology, 352(5), 1068–1080. [DOI] [PubMed] [Google Scholar]
- Williams AH, Wheeler R, Deghmane A-E, Santecchia I, Impens F, Bastos PAD, … Boneca IG (2019). Crippling the bacterial cell wall molecular machinery. BioRxiv, 607697 10.1101/607697 [DOI] [Google Scholar]
- Williams AH, Wheeler R, Rateau L, Malosse C, Chamot-Rooke J, Haouz A, … Boneca IG (2018). A step-by-step in crystallo guide to bond cleavage and 1,6-anhydro-sugar product synthesis by a peptidoglycan-degrading lytic transglycosylase. Journal of Biological Chemistry, 293(16), 6000–6010. 10.1074/jbc.RA117.001095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA, & Weiss DS (2015). Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proceedings of the National Academy of Sciences, 112(36), 11347–11352. 10.1073/pnas.1508536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, & Bernhardt TG (2011). An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proceedings of the National Academy of Sciences, 108(45), 18209 LP – 18210. Retrieved from http://www.pnas.org/content/108/45/E1052/1.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, & Xiao J (2017). GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science, 355(6326), 744 LP – 747. 10.1126/science.aak9995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunck R, Cho H, & Bernhardt TG (2016). Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Molecular Microbiology, 99(4), 700–718. 10.1111/mmi.13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Patel V, Helmann JD, & Dörr T (2017). Don’t let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Molecular Microbiology, 106(6), 847–860. 10.1111/mmi.13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.