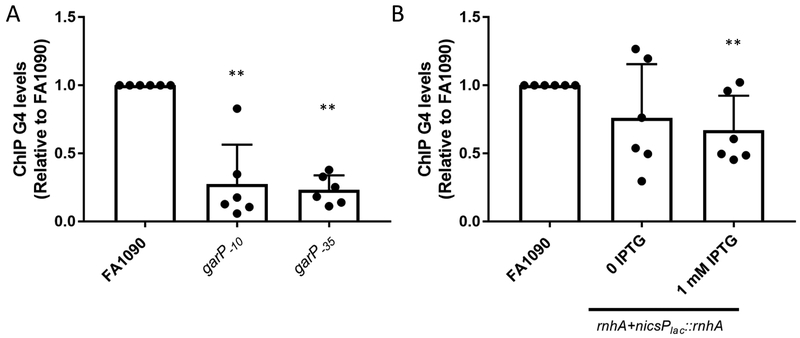
Figure 4.
Transcription initiation, but not R-loop stability, influences G4 levels
A. G4 levels were determined using G4 ChIP on garP−10 and garP−35 with the G4 specific MAb, 1H6). Relative G4 levels are calculated by determining the % input after ChIP pulldown for each sample and the dividing by the % input of FA1090. Bar The reported value represents the mean of six biological replicates (individual dots) +/− one standard deviation as indicated by the error bars. Unpaired student t-test **p<0.01
B. G4 ChIP was used to determine G4 levels under low rnhA and high rnhA expression (rnhA+nicsPlac::rnhA 0 IPTG, and rnhA+nicsPlac::rnhA 1 mM IPTG). The reported value represents the mean of six biological replicates (individual dots) +/− one standard deviation as indicated by the error bars. Unpaired student t-test**p<0.01