Figure 3.
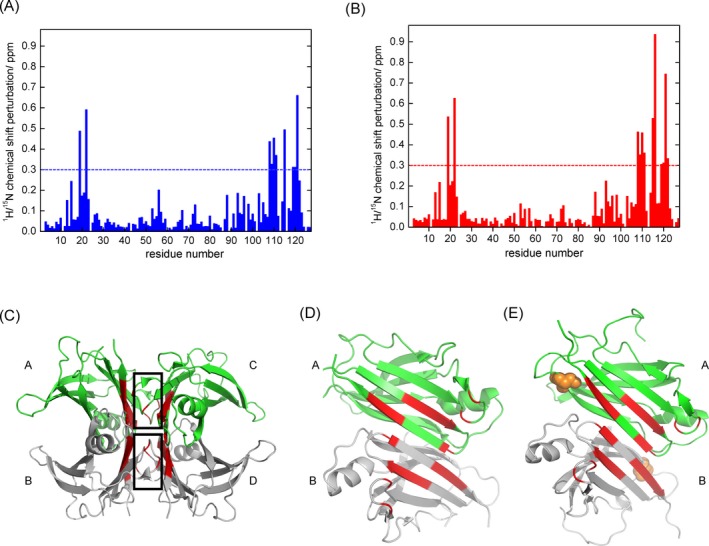
Characterizations of tafamidis binding sites of wildtype transthyretin and A97S transthyretin. (A) Normalized chemical shift changes for each residue of wildtype TTR due to tafamidis binding; (B) normalized chemical shift changes for each residue of A97S‐TTR due to tafamidis binding; (C) mapping of significantly perturbed (>0.3 ppm) residues in red color on the ribbon representation of the wildtype transthyretin using crystal structure (PBD ID: 4pvl), where the thyroxine binding sites are indicated with black box; (D) mapping of significantly perturbed (>0.3 ppm) residues on the ribbon representation of the dimer‐dimer interface of the wildtype transthyretin using crystal structure (PBD ID: 4pvl); (E) mapping of significantly perturbed (>0.3 ppm) residues on the ribbon representation of the dimer‐dimer interface of A97S transthyretin using a homology model generated based on the wildtype transthyretin crystal structure (PBD ID: 4pvl), where Ser‐97 residues are highlighted as orange spheres. In Figure 3D and E, the structural models were turned 90 degrees, with subunits C and B removed, to clearly show the TTR dimer‐dimer interface of subunit A and B.
