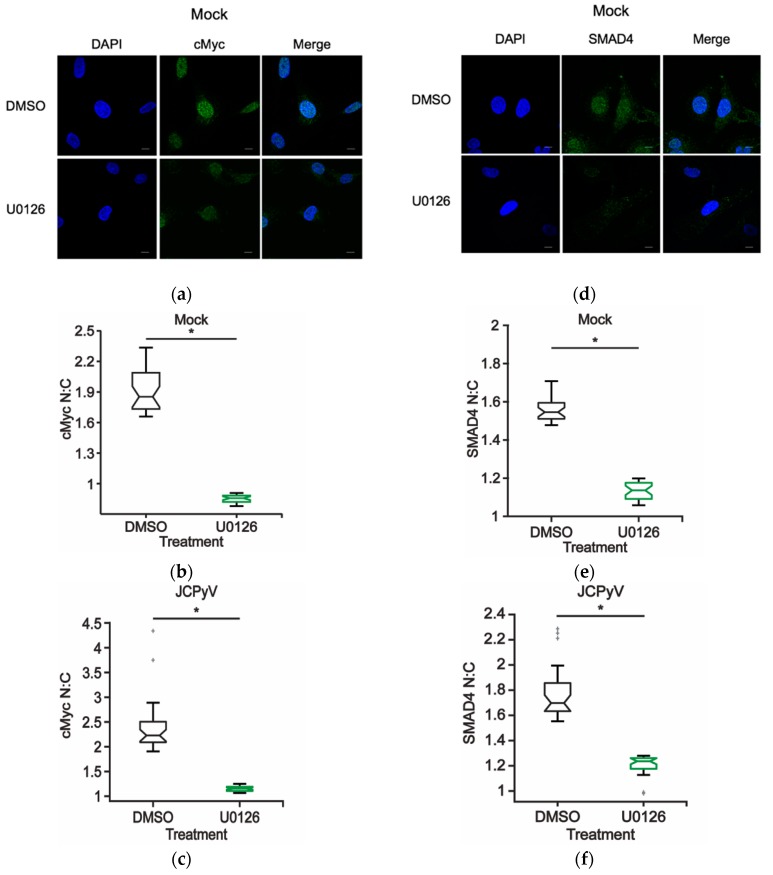
Figure 5.
ERK inhibition reduces nuclear localization of MAPK-associated transcription factors during JCPyV infection. SVG-A cells were treated with U0126 (10 μM) or DMSO (vehicle control) in cMEM and incubated at 37 °C for 48 h, then fixed and processed for confocal imaging. Representative confocal images demonstrating (a) cMyc and (d) SMAD4 (green) cellular localization patterns during DMSO- or U0126- treatment (10 μM) in mock-infected cells (100x). Cell nuclei stained with DAPI (blue). Scale bar = 10 μM. Boxplots of N:C ratios of (b) cMyc and (e) SMAD4 quantified from confocal images (60x) of 30 individual cells of DMSO- and U0126-treated mock-infected cells (60x). (c,f) SVG-A cells were treated with either U0126 (10 μM) or DMSO (vehicle control) containing cMEM for 1 h, infected with JCPyV (MOI: 2 FFU/cell), and incubated in cMEM at 37 °C for 1 h. At 1 hpi infected cells were fed with appropriately treated media (DMSO or U0126), incubated at 37 °C for 48 h, then fixed and processed for confocal imaging. Boxplots of N:C ratios for (c) cMyc and (f) SMAD4 quantified from confocal images of 30 individual cells of DMSO- and U0126-treated cells infected with JCPyV for 48 h. Whiskers represent values 1.5 times the distance between the first and third quartiles (inter-quartile range). Boxplot notches denote the 95% confidence interval around the median. Data are representative of N:C ratios from three independent experiments. Wilcoxon sum rank test was used to determine statistical significance. *, p < 0.05.