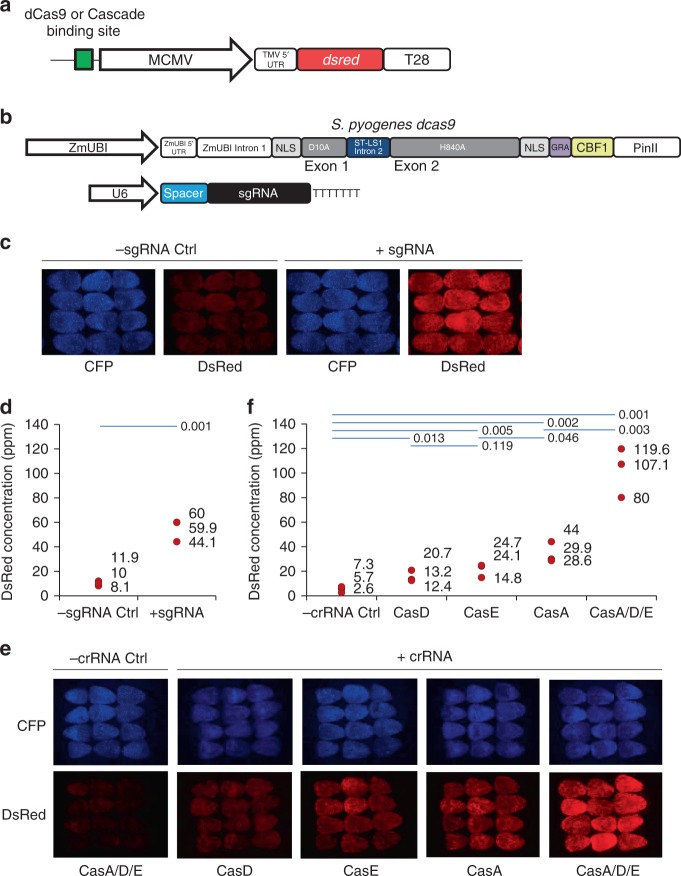
Fig. 2.
dCas9 and SthCascade dsred gene activation in Zea mays cells. a Expression construct for assaying CRISPR-induced gene activation in Zea mays. A single dCas9 or SthCascade binding site upstream of a minimal Cauliflower Mosaic Virus (MCMV) promoter, a Tobacco Mosaic Virus (TMV) 5′ UTR, and a maize codon conditioned gene encoding a variant of the reef coral red fluorescent protein from Discosoma sp. (DsRed) followed by a T28 terminator from Oryza sativa. b S. pyogenes dCas9-CBF1 transcriptional activation and sgRNA expression constructs for maize. c Confocal immunofluorescence images of dCas9-CBF1 particle gun transformed Zea mays immature embryos (IEs). Expression constructs encoding the cyan fluorescent protein (CFP) were co-delivered with other components as a control for transformation efficiency. Experiments assembled without a sgRNA expression cassette served as a negative control (-sgRNA Ctrl). d DsRed concentration in dCas9-CBF1 transformed IEs 48 h post transformation. Results from three independent transformations are graphed. Statistical significance between treatments (blue line) was evaluated using a one-sided t-test resulting in a p-value of 0.001. e CFP and DsRed images of SthCascade-CBF1 particle gun transformed IEs. CFP fluorescence served as a control for transformation efficiency. Experiments performed with CasA-CBF1, CasB, CasC, CasD-CBF1, and CasE-CBF1 expression cassettes in the absence of a crRNA expression construct served as the negative control (-crRNA Ctrl). f Quantification of DsRed protein resulting from SthCascade transcriptional activation 48 h after transformation in three independent experiments. The p-values resulting from a one-sided t-test between the indicated treatments (blue lines) are shown