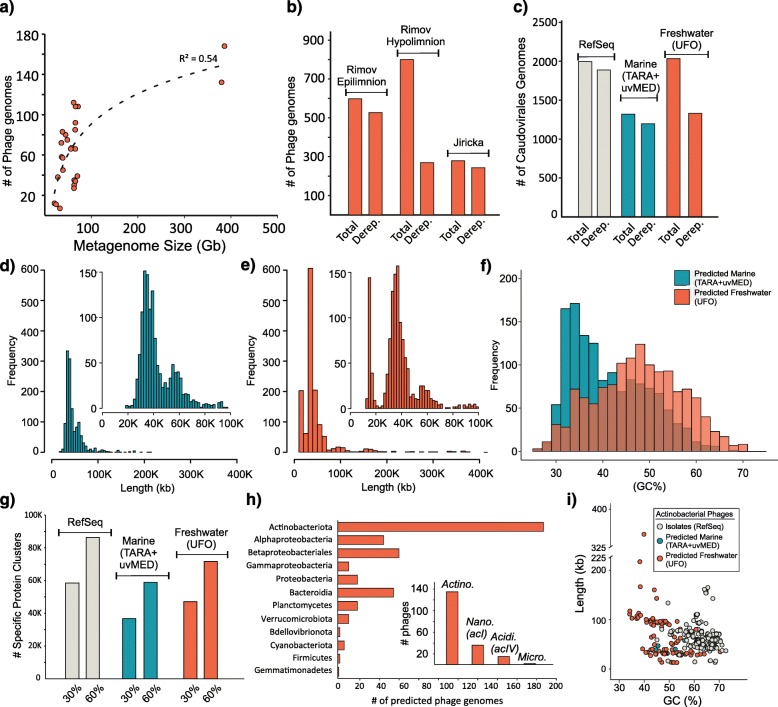
Fig. 1.
Genome statistics. a Complete phage genome recovery as a function of metagenome size. b Total number of phage genomes vs dereplicated genomes from Řimov reservoir (epi- and hypolimnion) and Jiřicka Pond. c Comparison of all phage genomes and dereplicated phage genomes from Viral Refseq, Marine habitat (TARA+uvMED) and the freshwater habitat (UFO). d Length distribution of marine phage genomes (inset, enlarged view of phages up to length 100 K). e Length distribution of recovered freshwater phage genomes (inset, enlarged view of phages up to length 100 K). f Comparative GC% distributions of marine and freshwater phage genomes. g Number of unique protein clusters in RefSeq, Marine, and Freshwater phage genomes at 30% and 60% identity. h Number of phage genomes with predicted host of the freshwater dataset (inset, the same for lower taxonomic ranks within Actinobacteria; Actino, Actinobacteriota; Nano, Nanopelagicales; Acidi, Acidimicrobiales; Micro, Microbacteriales). i GC% vs length for predicted (freshwater, marine) and known actinobacterial phages (RefSeq)