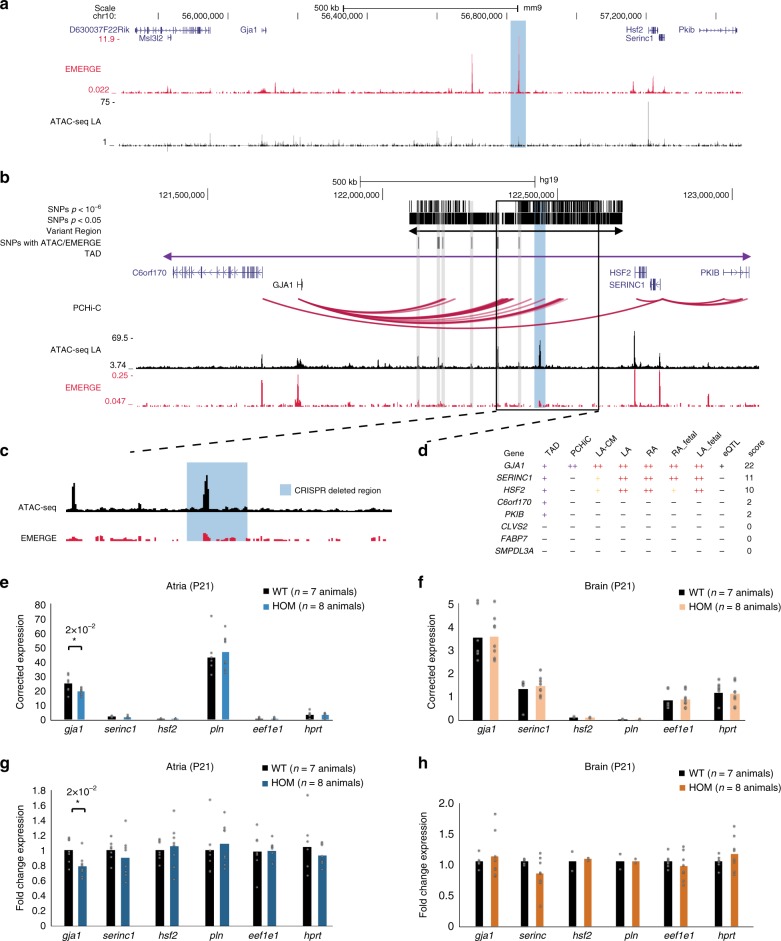
Fig. 5.
Variant region deletion shows distal regulatory element downstream of Gja1 in mouse. a Mouse region homologous to human AF associated GJA1 locus, showing gene, EMERGE and ATAC-seq tracks. Highlighted region is the 40 Kb deletion. b Browser view of human GJA1 locus, showing AF associated (subthreshold) SNPs in variant region (VR), TAD, genes, cardiac PCHi-C, ATAC-seq and EMERGE. Human homologous deleted region (highlighted). c Zoom of the human homologous deleted region (highlighted.) d Expression and PCHi-C values per gene in the TAD. e Corrected expression of genes as assessed by qPCR with Hprt and Eef1e1 as reference genes in atria and f brain. g Expression fold change of Gja1RE1/RE1 (HOM) vs wild-type (WT) littermates as a negative control of P21 atria and h brain cortex. Source data of 5e–h are provided as Source Data files. Error bars represent SD, *p < 0.05, **p < 0.01, and ***p < 0.001 (two-tailed Student’s t-test). kb kilobase, SNP single nucleotide polymorphism, TAD topologically associated domain, PCHi-C promoter capture Hi-C, ATAC-seq assay for transposase-accessible chromatin sequencing, qPCR quantitative polymerase chain reaction, P21 postnatal day 21, SD standard deviation