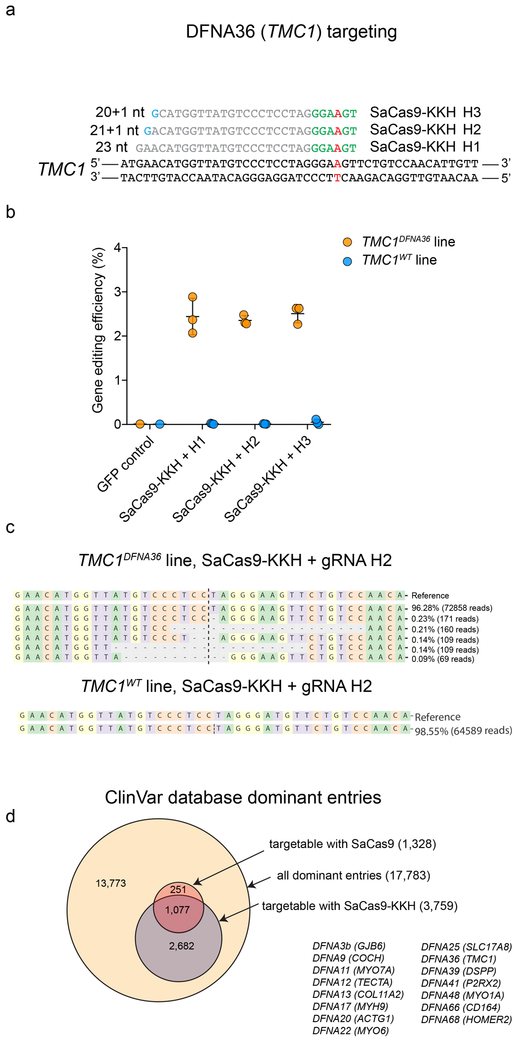
Figure 4:
Human dominant mutations, potentially targetable with SaCas9 and SaCas9-KKH, and allele-specific targeting of human DFNA36. (a) All human dominant mutations in the ClinVar database (accessed 2019.03.25) and mutations targetable with SaCas9 and SaCas9-KKH. (b) gRNA design targeting a human DFNA36 allele. Mutation site is highlighted in red. PAM sites are marked by green nucleotides. Mismatching nucleotides are shown in blue. (c) Genome editing efficiency (indel formation percentage) in haploid TMC1DFNA36 and TMC1WT cells after transfection with SaCas9-KKH and H1, H2, H3 gRNA (mean ± SD, 3 biological replicates sequenced independently). Control cells were transfected with GFP only. (d) Types of indels in the case of H2 gRNA in TMC1DFNA36 and TMC1WT cells (CRISPResso analysis, similar results were obtained from all gRNAs and all biological replicates).