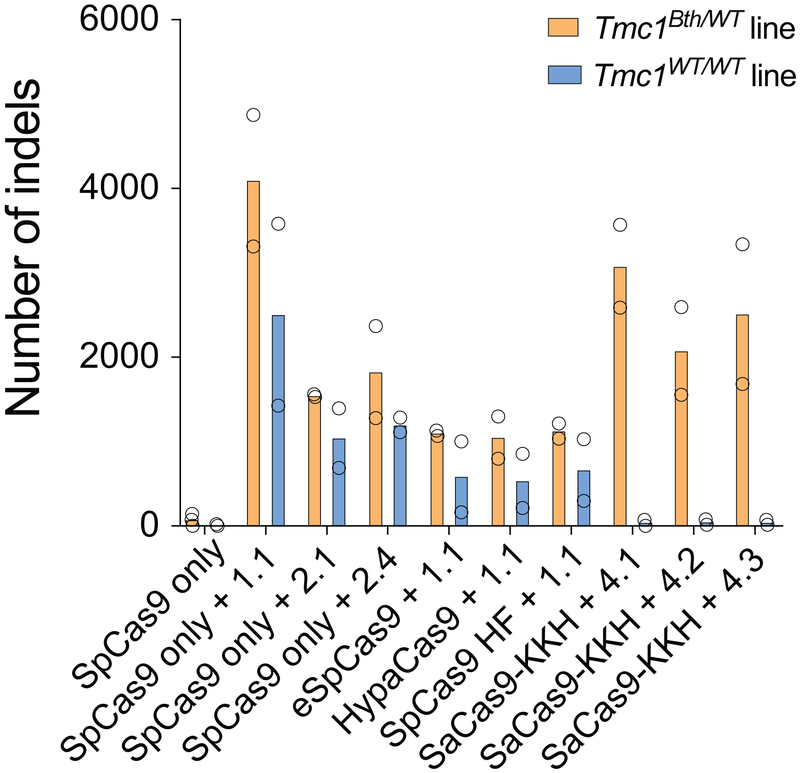
Extended Data Fig. 2.
Number of indels based on targeted deep sequencing data from Tmc1Bth/WT and Tmc1WT/WT cell lines treated with different Cas9+gRNA combinations (from Fig. 1b). Note, that data points show non-normalized read counts. Cells were transfected on two different occasions (SpCas9 only and SpCas9 + gRNA 1.1 on four occasions) and genomic DNA from two independent biological samples on each transfection day were pooled for sequencing. Indels in the SaCas9-KKH treated Tmc1WT/WT lines are not different from the background (i.e. untreated samples). This method revealed high sensitivity, as the indel rates in CRISPR treated samples were 40-160-fold higher than the background indel rates observed in untreated samples.