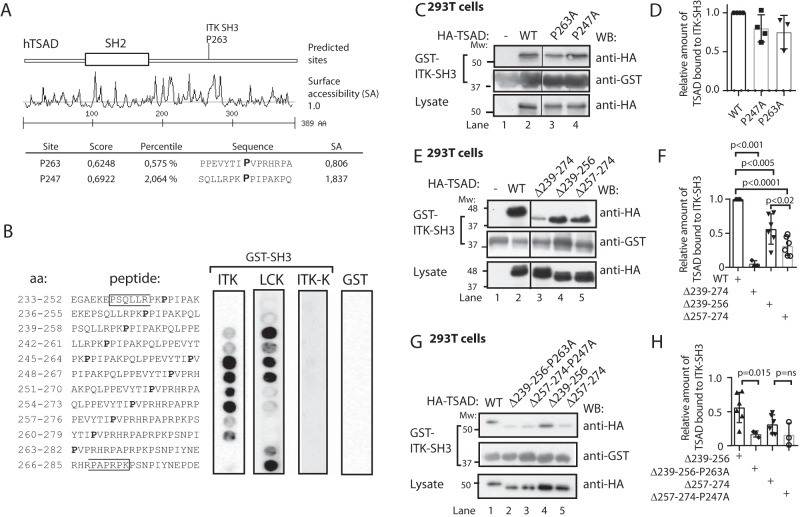
Figure 2.
LCK SH3 and ITK SH3 bind to overlapping peptides in TSAD PRR. A, Scansite prediction of ITK SH3-binding sites on TSAD. B, peptide array mapping of LCK SH3- and ITK SH3-binding sites on TSAD. The amino acid sequence of the peptide in each of the spots is given to the left. The right panels show the results of probing the arrays with the indicated recombinant proteins GST-ITK SH3, GST-LCK SH3, GST-ITK SH3 mutated (W208K or ITK-K), or GST alone, respectively. Results from 12 of the 14 tested peptides are shown. Pro247 and Pro263 are indicated in boldface type. The brackets define the borders of TSAD aa 239–274. C and D, mutations of the predicted prolines to alanines fail to abolish binding of ITK SH3 to TSAD. The pulldown experiment was performed using GST-ITK SH3 and lysates of 293T cells transiently transfected with HA-tagged WT and mutated TSAD cDNA. Pulled down protein and lysate were immunoblotted with the indicated antibodies. D, the graph represents the relative amount of TSAD interacting with ITK SH3 in the experiment shown in C. Signals were quantified by ImageJ analysis (n = 3, mean ± S.D. (error bars)). E and F, deletion of either the Pro247 (aa 239–256) or the Pro263 (aa 257–274) region reduces binding of ITK SH3 to TSAD. The pulldown experiment was performed as in C. F, the graph represents ImageJ analysis as in D of the data shown in E (n = 3 or 6, mean ± S.D.). G and H, additional mutation of Pro263 further reduces binding of ITK SH3 to TSAD 257–274. The pulldown experiment was performed as in C. H, the graph represents ImageJ analysis as in D of data shown in G. The relative amount of TSAD interacting with ITK SH3 WT in each experiment is set to 1 (not shown) (n = 3, mean ± S.D.).