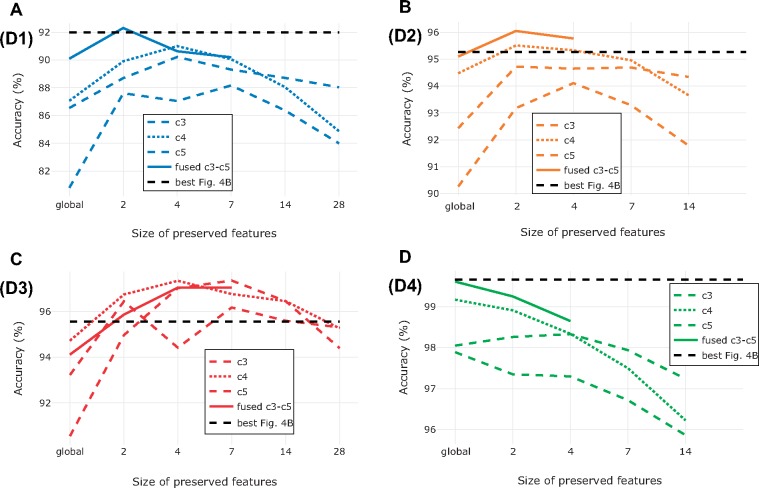
Figure 5.
Impact of preserving more features during feature extraction on identification accuracy for data sets D1 (a), D2 (b), D3 (c), and D4 (d). We used input images of size 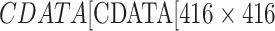 in all cases, and we pooled features from each of the last three convolutional blocks (c3–c5) into matrices of dimension
in all cases, and we pooled features from each of the last three convolutional blocks (c3–c5) into matrices of dimension 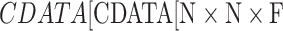 , where F is the number of filters of the corresponding convolutional block and N was set to 1 (global average pooling), 2, 4, 7, 14, or 28. The dashed black line indicates the best performing model from previous experiments (Fig. 4b). For D2 and D4 we stopped at N = 14 because of the computational cost involved in computing the value for N = 28, and since the accuracy had already dropped significantly at N = 14. Because of the computational cost, we computed performance of fused feature matrices only up to N = 7 (D1 and D3) or N = 4 (D2 and D4). On data sets D1, D2, and D3, the optimum identification performance was somewhere between globally pooled and nonpooled features. For D4, global pooling yielded the best identification performance.
, where F is the number of filters of the corresponding convolutional block and N was set to 1 (global average pooling), 2, 4, 7, 14, or 28. The dashed black line indicates the best performing model from previous experiments (Fig. 4b). For D2 and D4 we stopped at N = 14 because of the computational cost involved in computing the value for N = 28, and since the accuracy had already dropped significantly at N = 14. Because of the computational cost, we computed performance of fused feature matrices only up to N = 7 (D1 and D3) or N = 4 (D2 and D4). On data sets D1, D2, and D3, the optimum identification performance was somewhere between globally pooled and nonpooled features. For D4, global pooling yielded the best identification performance.