Table 3.
Experiments on recently published biological image classification tasks
| Data sets | Categories | Our method | Original study | Method | Reference |
|---|---|---|---|---|---|
| ClearedLeaf | 19 families | 88.7 | 71 | SIFT + SVM | (Wilf et al., 2016) |
| CRLeaves | 255 species | 94.67 | 51 | finetune InceptionV3 | (Carranza-Rojas et al., 2017) |
| EcuadorMoths | 675 species | 55.4 (58.2) |
55.7 57.19 |
AlexNet + SVM VGG16 + SCDA + SVM |
(Rodner et al., 2015) (Wei et al., 2017) |
| Flavia | 32 species | 99.95 | 99.65 | ResNet26 | (Sun et al., 2017) |
| Pollen23 | 23 species | 94.8 | 64 | CST + BOW | (Gonçalves et al., 2016) |
Note: We used input images of size 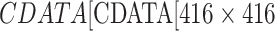 (
(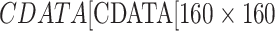 for Pollen23), global average pooling, fusion of c1–c5 features, and signed square root normalization. Accuracy is reported using 10-fold cross validation except for EcuadorMoths, where we used the same partitioning as in the original study. Bold font indicates the best identification performance.
for Pollen23), global average pooling, fusion of c1–c5 features, and signed square root normalization. Accuracy is reported using 10-fold cross validation except for EcuadorMoths, where we used the same partitioning as in the original study. Bold font indicates the best identification performance.
