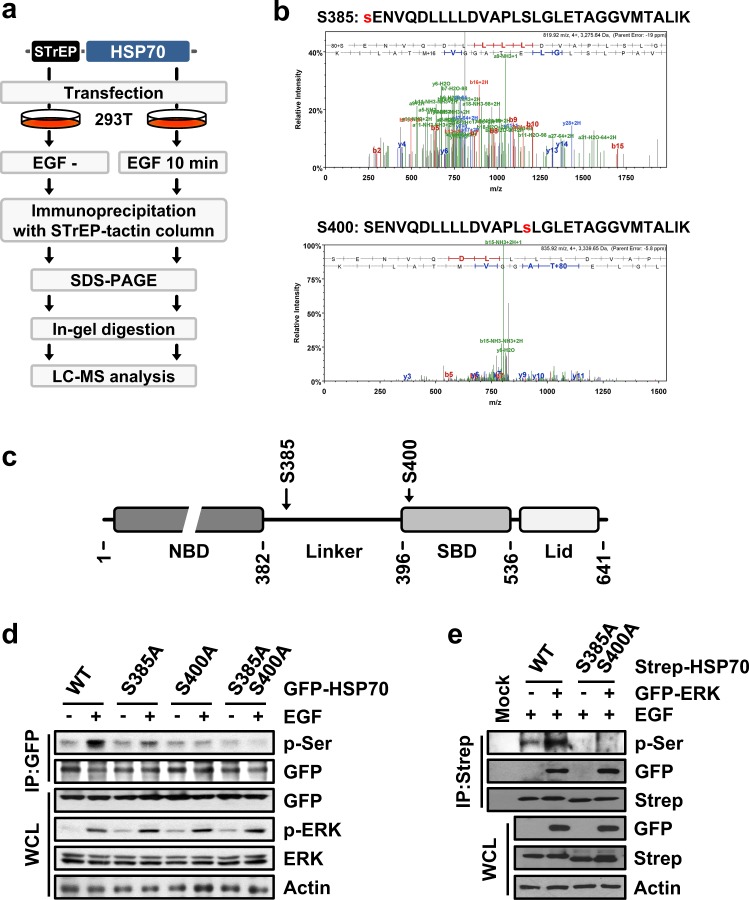
Fig. 2. Identification of HSP70 phosphorylation residues.
a, b LC-MS analysis for the identification of the phosphorylated residues of HSP70 a Strategy used for the identification of HSP70 phosphorylation sites via LC-MS analysis. b Mass spectrum indicating the phosphorylation of HSP70 serine residues at positions 385 (upper) and 400 (bottom). The identified peptide sequence, including the phosphorylated residue, is presented above its spectrum. Phosphorylated serines are colored red. c Cartoon showing HSP70 domain arrangement. NBD and SBD indicate the nucleotide-binding domain and the substrate-binding domain, respectively. Serine residues at positions 385 and 400 were located in the linker and SBD regions, respectively. d The indicated mutants-expressing 293T cells were either treated or not treated with EGF, and phosphorylation was determined by immunoblotting using specific antibodies. e Strep-HSP70 and GFP-ERK were introduced into 293T cells as indicated. HSP70 phosphorylation and binding to ERK were detected by Western blotting