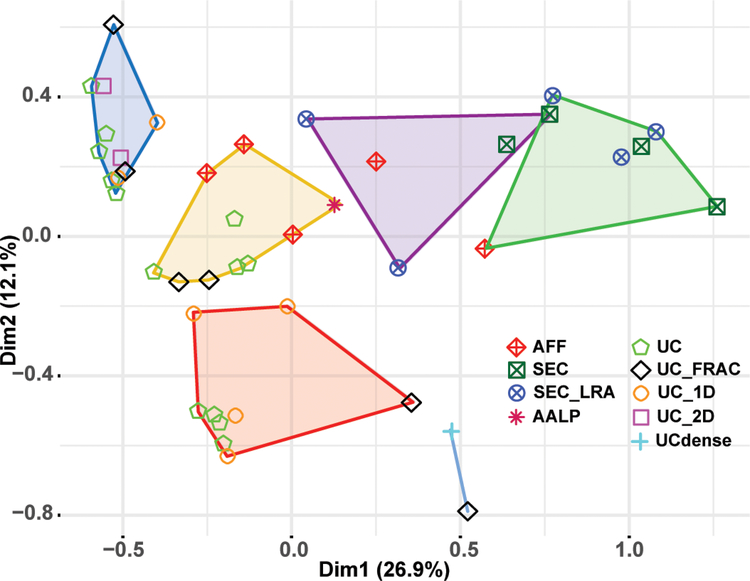
Figure 2.
Multiple correspondence analysis (MCA) and hierarchical clustering of individual studies. MCA was performed with a subset of proteins that were identified in at least 5 different studies (n = 139). Individual studies were grouped by hierarchical clustering based on the eigenvalues for each study obtained from the MCA (e.g. based on similarity of the protein ensemble identified). The clusters of closely related studies are represented by colored polygons. Each study is further denoted by an individual symbol corresponding to the HDL isolation methodology used.