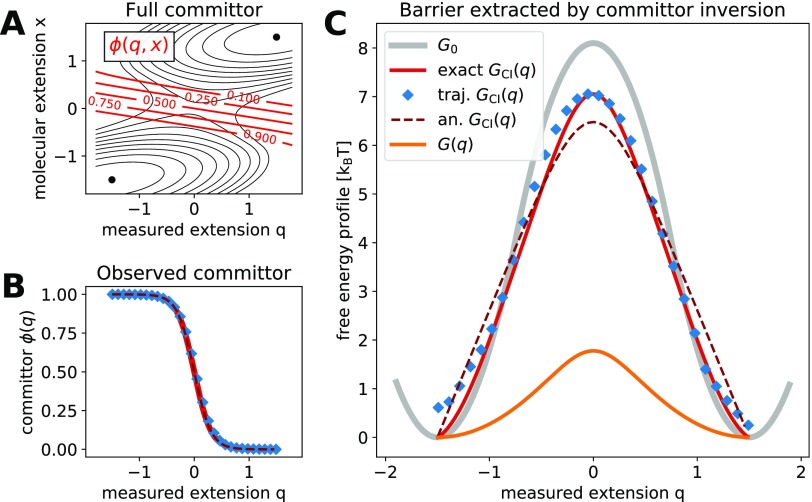
FIG. 3.
Molecular free energy profile from committor inversion. (a) Full two-dimensional committor for the free energy surface G(q, x) and Dq/Dx = 1. Isolines of the free energy surface are shown as black solid lines separated by 1 kBT. Isolines of the committor are shown as red solid lines. (b) The observed committor ϕ(q) is obtained in two independent ways: from a numerical solution of Onsager’s equation (exact, red solid line) and from Brownian dynamics trajectories (blue diamonds). The analytic prediction from Eq. (9) is shown as a dark red dashed line. (c) The free energy barrier extracted by committor inversion, GCI(q), from the exact and trajectory-estimated committor (red solid line and blue diamonds, respectively). Both barriers are compared to the hidden molecular profile Go (gray solid line), to the analytic prediction from Eq. (11) (dark red dashed line), and to the Boltzmann-inverted free energy profile G(q) (orange solid line). The free energy surface G(q, x) has parameters similar to those obtained for the 20TS06/T4 DNA hairpin,19 , Δx‡ = 1.5 [x], and κl = 2.6 kBT/[q]2, where [q] = [x] denotes units of length for the extension.