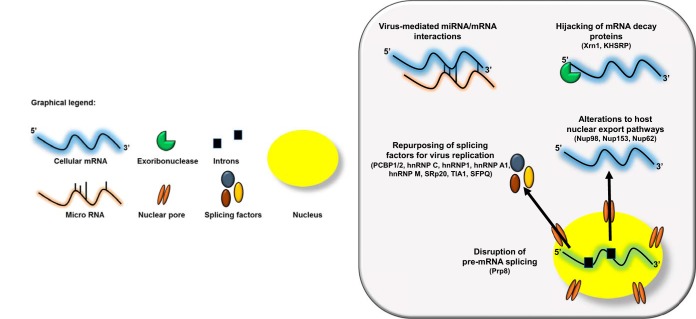
FIG 1.
Picornaviruses alter host RNA metabolism at multiple steps. The viral-RNA-dependent RNA polymerase 3Dpol of the picornavirus EV71 disrupts host cell pre-mRNA splicing via its interaction with the core splicing machinery protein Prp8. Nuclear cytoplasmic trafficking is also dysregulated during picornavirus infection via either the cleavage of nuclear pore complexes (NPCs) by the enteroviral proteinase 2Apro or hyperphosphorylation of the NPC by cardiovirus L protein. These alterations result in the nuclear retention of certain classes of cellular RNAs, in addition to the cytoplasmic relocalization of a suite of splicing factors required for picornavirus replication. Several picornaviruses hijack mRNA decay proteins, most notably the processing body protein Xrn1, to facilitate their replication cycle as well as an interaction that positively regulates autophagy during picornavirus infection and provides further evidence for the AWOL hypothesis for picornavirus egress. Finally, picornaviruses have been shown to mediate several novel cellular mRNA-miRNA interactions, which have implications for viral pathogenesis.