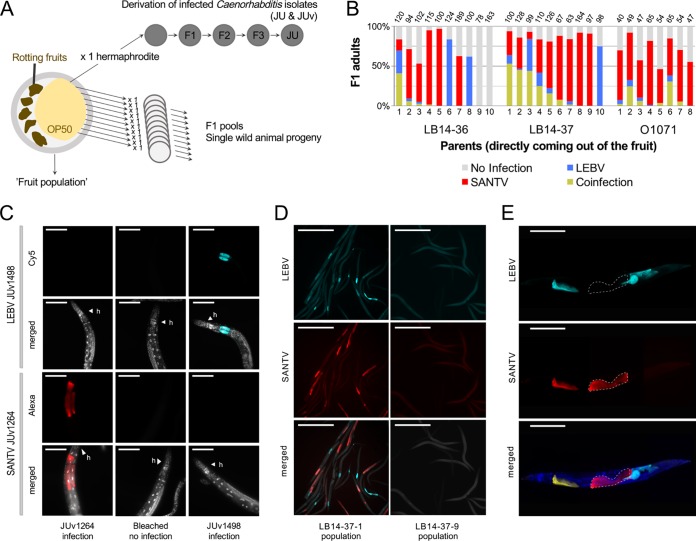
FIG 3.
Coinfection of C. briggsae by Santeuil and Le Blanc viruses at the level of population, individual animals, and single cells. (A) Design of the assay. Rotten vegetal matter was collected from the wild, brought back to the laboratory, and plated on an E. coli OP50-seeded NGM plate. Single hermaphrodites of C. briggsae were isolated onto a new plate. C. briggsae wild isolates (JU) naturally infected with their noda-like viruses (JUv) were derived from the fruit population. In the survey for coinfection (shown in panel B), subsets of the C. briggsae wild populations coming out of the fruit (fruit population) were fixed in ethanol, and the presence of viruses was systematically monitored using in situ hybridization. (B) Natural coinfection in the progeny of wild animals (numbered on the x axis) isolated from rotten fruits. Three samples where SANTV and LEBV were present were chosen to assess the coexistence of the two viruses at the level of individual worms: a plum (LB14-36) and a pear (LB14-37) from the Le Blanc location and an apple (O1071) from the Orsay orchard. The F1 progeny (F1 pool) of 8 to 10 parents isolated from the parental populations were fixed in ethanol when they reached the adult stage. Many of the isolated adults harbored both viruses as assayed by their presence in their progeny, and some of their progeny also detectably harbored both viruses. Note that in all three cases the Santeuil virus appears predominant. The number of scored animals in each F1 pool is indicated on top of the bar. (C) Specificity of the ssDNA probes used to detect LEBV and SANTV. The JU1264 C. briggsae strain was experimentally infected with SANTV JUv1264 and with LEBV JUv1498 in the laboratory. Uninfected JU1264 C. briggsae animals were used as a control. Shown is fluorescence in situ hybridization staining of SANTV- and LEBV-infected animals using SANTV RNA1 (Cal Fluor red 610, 40×, 200 ms) and LEBV RNA1 (Quasar 670, 40×, 200 ms) Biosearch probes (see Table S1b). Nuclei were counterstained with DAPI (merged panels). Red, SANTV RNA1 probe; cyan, LEBV RNA1 probe; gray, DAPI. Scale bars represent 100 μm. (D) Coinfection at the level of the single C. briggsae LB14-37.1 F1 population. The uninfected C. briggsae F1 population LB14-37.9 was chosen as a control. Shown is fluorescence in situ hybridization staining of SANTV- and LEBV-infected animals using SANTV RNA1 (Cal Fluor red 610) and LEBV RNA1 (Quasar 670) probe sets (see Table S1b). Nuclei were counterstained with DAPI (merged panels). Red, SANTV RNA1 probe; cyan, LEBV RNA1 probe. Scale bars represent 500 μm. (E) Coinfections were recorded at the level of a single individual and a single cell both in naturally and in experimentally infected C. briggsae animals. Here, the illustration is of a C. briggsae adult hermaphrodite (strain JU1264) experimentally coinfected with JUv1264 and JUv1498 in the laboratory. Shown is fluorescence in situ hybridization staining of SANTV- and LEBV-infected animals using SANTV RNA1 (Cal Fluor red 610) and LEBV RNA1 (Quasar 670) probe sets (see Table S1b). Nuclei were counterstained with DAPI (merged panels). Dashed white lines delineate two intestinal cells infected with JUv1264 virus and not with JUv1498, exemplifying the probe specificity to each virus. Red, SANTV RNA1 probe; cyan, LEBV RNA1 probe; yellow, coinfected cells; blue, DAPI. Scale bars represent 100 μm.