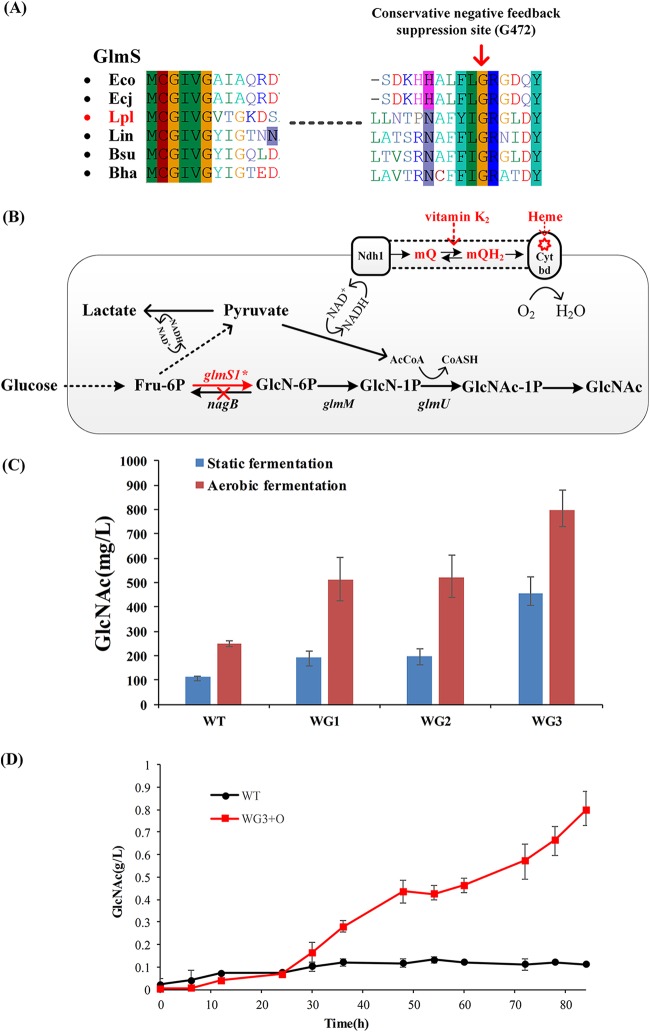
FIG 5.
Increase of GlcNAc production by strain and process optimization. (A) Amino acid sequence comparison of GlmS to six other bacterial strains. In E. coli, G471S was proved to effectively relieve product resistance. By comparison of the amino acid sequences of glmS, G471 is highly conserved in these alignments. Therefore, we chose this conserved site (G472 in Lactobacillus plantarum WCFS1) mutation to conduct recombineering using a designed ssDNA. The G472 site is indicated by a red arrow. Organism abbreviations: Eco, E. coli K-12 MG1655; Ecj, E. coli K-12 W3110; Lpl, L. plantarum WCFS1; Lin, Listeria innocua (serotype 6a); Bsu, Bacillus subtilis subsp. subtilis 168; Bha, Bacillus halodurans. (B) Optimized metabolic pathways for GlcNAc production. Optimization was accomplished via mutation of glmS1 and knockout of nagB. Heme and menaquinone were external sources and are shown in red. (C) Fermentation production of GlcNAc in each individual engineered strain. Aerobic fermentation was conducted in MRS medium at 37°C and 220 rpm, and pH was maintained at approximately 6.5. Results are averages from three independent experiments. (D) Fermentation profiles of wild-type (WT) L. plantarum WCFS1 under anaerobic conditions and WG3 under aerobic conditions.