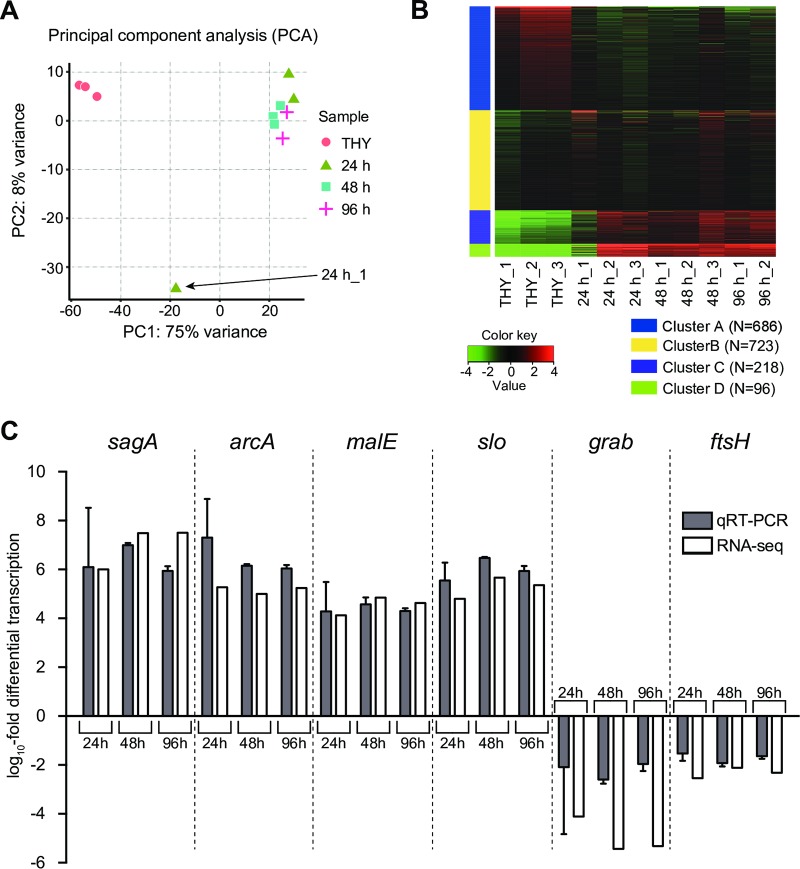
FIG 2.
RNA-seq global reports. (A) Principal component analysis (PCA) plot of RPKM data from RNA-seq data set. (B) Heatmap of k means clustering of all genes (1,723 genes) in all samples (k = 4). The number of expressed genes in each cluster is indicated. The color key indicates Z score and displays the relative values of all tiles within all samples: green, lowest expression; black, intermediate expression; red, highest expression. Bacterial RNA-seq data at 24, 48, and 96 h postinfection were defined as the 24-h (24 h_1, 24 h_2, and 24 h_3), 48-h (48 h_1, 48 h_2, and 48 h_3), and 96-h (96 h_1 and 96 h_2) groups, respectively. Bacterial RNA-seq data of THY culture samples were defined as the control, termed the THY group (THY_1, THY_2, and THY_3). (C) Validation of RNA-seq data using qRT-PCR. The x axis shows selected genes subjected to qRT-PCR assays, and the y axis shows the log10 fold change relative to that of THY culture samples. qRT-PCR was performed with total RNA from the samples used for RNA-seq. Data from two or three independent qRT-PCR assays, each performed in triplicate, were pooled and normalized. rpoB was used as the internal control. Vertical lines represent the means ± standard errors (SE).