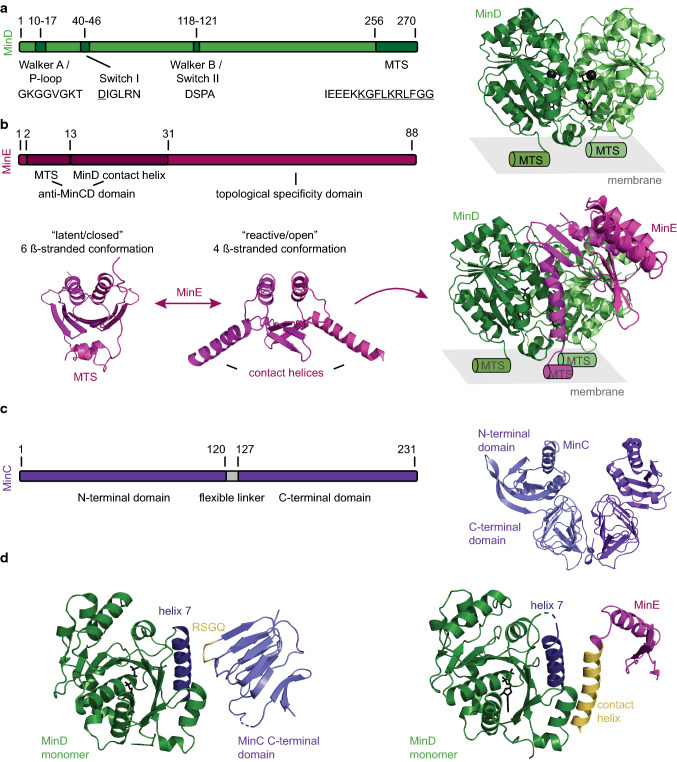
Fig. 1.
Overview of the three Min proteins. a MinD can bind to the membrane via its C-terminal MTS upon ATP-dependent dimerization (residues forming the amphipathic helix are underlined). Schematic view highlighting the structural motifs and their respective amino acid sequences of E. coli MinD: Walker A and B and switch I motifs required for ATP binding and Mg2+ coordination. Crystal structure of the dimeric MinD with ATP and the Mg2+ ion shown in black. MTS location is indicated by schematic helices (PDB: 3Q9L [36]). b Schematic view highlighting the structural motifs and their respective amino acid sequences of E. coli MinE. MinE exists in a latent/closed conformation in solution (crystal structure of Neisseria gonorrhoeae MinE, PDB:2KX0 [51]). Upon “sensing” MinD on the membrane, it transforms into a reactive/open conformation where the contact helices and MTS are exposed, freeing it to interact with MinD (crystal structure of the E. coli MinDE complex, note that MinE 13-88 I24N was used, PDB: 3R9J [52]). c Schematic view of E. coli MinC shows that the protein consists of two domains that are connected via a flexible linker. Crystal structure of dimeric MinC from Thermotoga maritima (PDB: 1HF2 [53]). d MinC and MinE have overlapping binding interfaces on MinD. Crystal structure of the Aquifex aeolicus MinD monomer with helix 7 highlighted in blue and the C-terminal domain of A. aeolicus MinC with the RSGQ motif displayed in yellow (left, PDB: 4V02 [54], note that helix 3 of A. aeolicus is not shown as it is absent in MinC of most bacterial species including E. coli [55]). Crystal structure of the E. coli MinD monomer with helix 7 highlighted in blue and MinE with the contact helix displayed in yellow (right, PDB: 3R9J [52], note that only the monomer of MinE is shown)