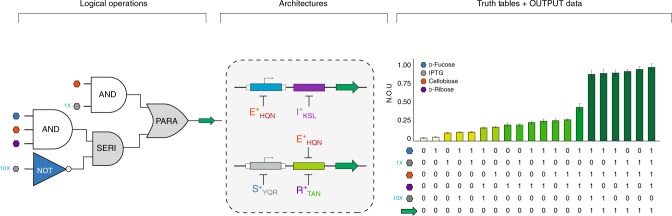
Fig. 8.
A complex program via the master architecture. Arranging two SERI structures in a PARA configuration leads to the development of the genetic master circuit. This configuration employs four non-synonymous DNA operators and allows for the generation of any of the logical programs presented in this study. When both outputs are the same (GFP), this gives rise to the most granular digital-to-analog converter in this study. The top channel contains two operators arranged in a SERI architecture and therefore functions as a digital-like AND gate with respect to cellobiose and IPTG. By capitalizing on off-diagonal interactions, the bottom channel becomes a three-signal AND gate with respect to the ligands d-fucose, d-ribose, and cellobiose. Finally, we take advantage of the ligand overlap between GalS and LacI to generate additional output states with varying GFP outputs. Values represent the mean of n = 12 biological replicates and error bars indicate the 95% CI or ~2x s.e.m. Y-axis is given as normalized output units or N.O.U. This is calculated by taking GFP fluorescence (485ex., 510em.) normalized to OD600, which is further normalized to the maximum output for each experiment. Source data provided as a Source Data File and individual data points overlaid as dot plots can be found in Supplementary Fig. 11