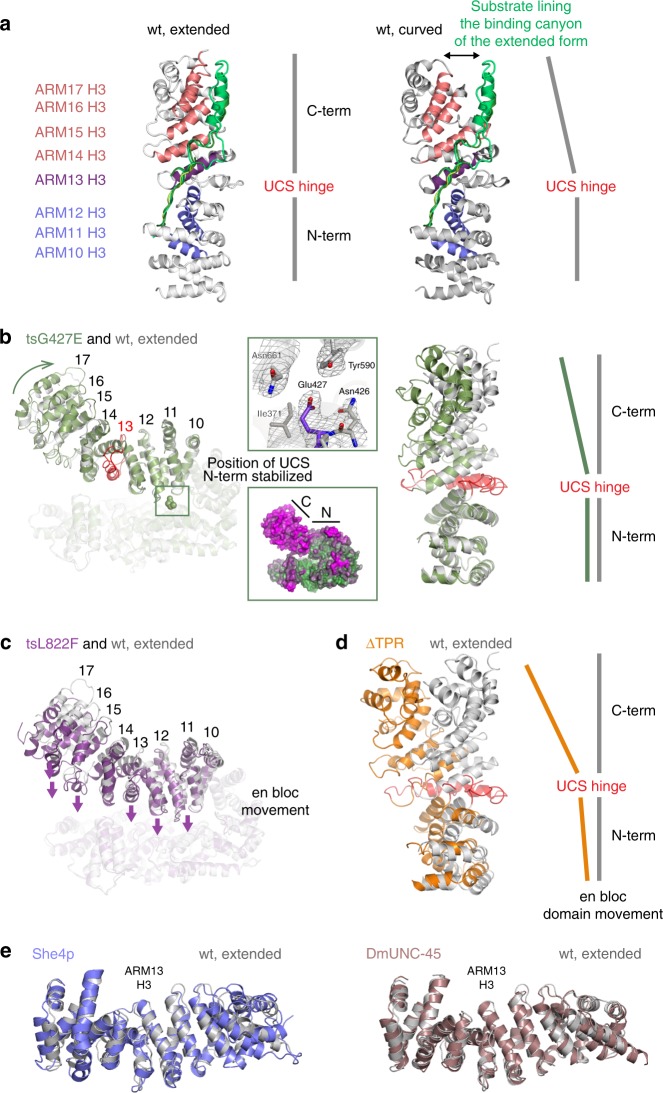
Fig. 5.
Structural analysis of UNC-45 mutant proteins. a Structural alignment of two different wild-type UNC-45 structures—PDB code: 4i2z, extended UCS domain conformation, PDB code 5mzu, curved UCS conformation. The three helices of each ARM repeat are organized such that helices H3 (colored) form the bottom of the myosin-binding canyon, whereas helices H1 and H2 compose the border of the molecular cleft. Substrates binding to the myosin binding-canyon are illustrated by overlay of ligands co-crystallized with the ARM domain of beta-catenin. b Structural alignment of wild-type UNC-45 (PDB code: 4i2z) and UNC-45 tsG427E—side view (left panel), top view (right panel). The top inset shows the E427 residue overlaid with 2fofc density contoured at 1σ. The bottom inset shows the thermal motion (b) factors displayed on the surface of the tsG427E mutant. N- and C-terminal subdomains of the UCS domain are indicated (green – rigid, magenta – flexible). c Structural alignment of wild-type UNC-45 (PDB code: 4i2z) and tsL822F. Arrows indicate the en bloc movement of the entire UCS domain towards the TPR/central domain. d Structural alignment of wild-type UNC-45 (PDB code: 4i2z) and UNC-45 ΔTPR molecule B. All structural alignments were performed with residues 1–523 as reference. e Structural alignment of wild-type UNC-45 (PDB code: 4i2z) and the UCS proteins She4p (PDB code: 3opb) and DmUNC-45 (PDB code: 3now). The center of the UCS myosin binding canyon (ARM 13 H3) is highlighted