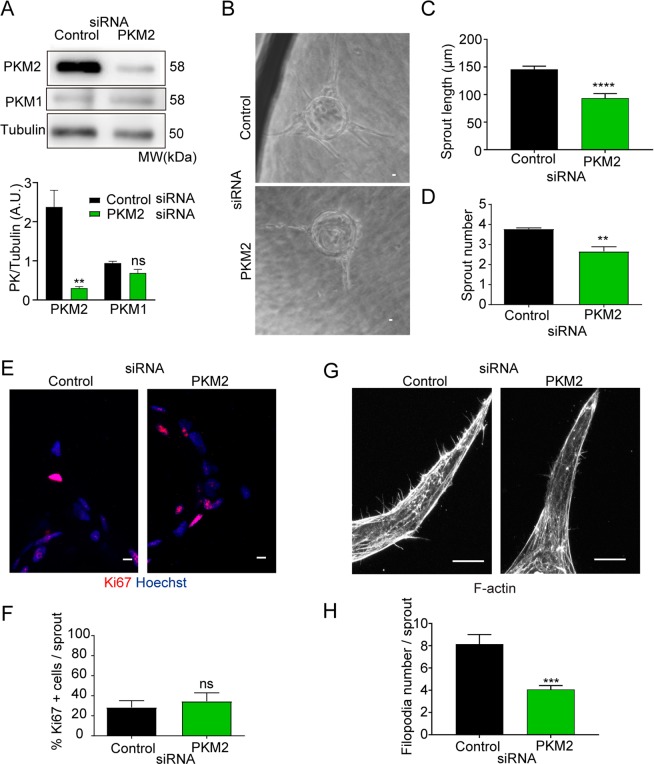
Figure 1.
PKM2 is required for in vitro endothelial cell sprouting. (A) Western blot of PKM2 and PKM1 expression 72 hours after siRNA silencing in HUVECs and quantification versus tubulin included as a loading control; means ± SEM, n = 3, ns non-significant, **p < 0.01 by unpaired Student t-test. (B) Bright-field microscopy images of spheroids coated with HUVECs transfected with control or PKM2 siRNA and embedded in fibrin gels for 7 days. Scale bar, 10 µm. (C) Sprout length in 3D spheroids; means ± SEM, n = 103 and 38 spheroids formed by control and PKM2 siRNA-silenced cells from one representative experiment of five performed, ****p < 0.0001 by unpaired Student t-test. (D) Sprout numbers in 3D spheroids; means ± SEM, n = 27 and 14 spheroids formed by control and PKM2 siRNA-silenced cells from one experiment representative of five performed, **p < 0.01 by unpaired Student t-test. (E) Immunofluorescence of Ki67 (red, proliferation) and Hoechst (blue, nuclei) in 3D spheroid sprouts. Scale bar, 10 µm. (F) Percentage of Ki67-positive cells per sprout in 3D spheroids; means ± SEM, n = 3 independent experiments, ns non-significant by paired Student t-test. (G) Immunofluorescence of F-actin in 3D spheroid sprouts. Scale bar, 10 µm. (H) Filopodia number in 3D spheroids; means ± SEM, n = 13 and 15 filopodia in sprouts formed by control and PKM2 siRNA-silenced cells from one representative experiment of five performed, ***p < 0.0001 by Welch´s test. MW, molecular weight. See also Figure S1.