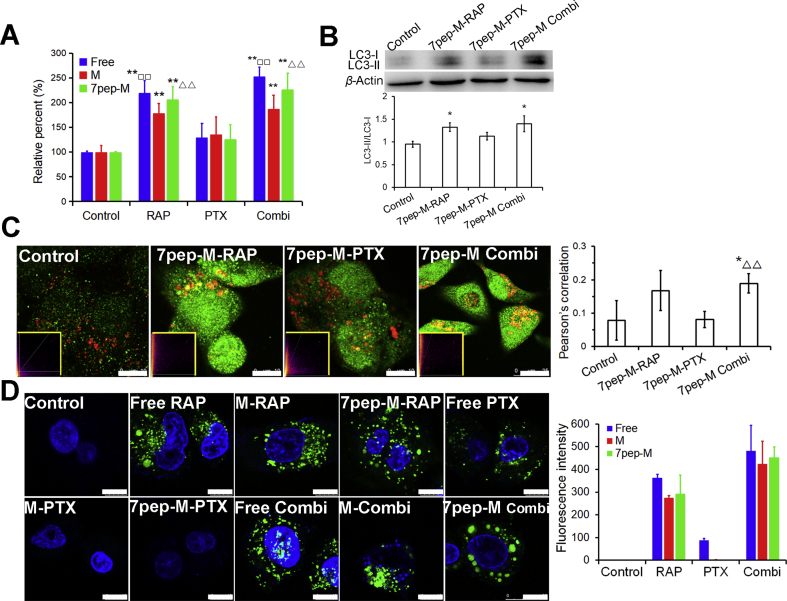
Figure 3.
Active autophagy modulation of 7pep-M-RAP. (A) Quantitative analysis of cellular LC3B by ELISA assay (mean ± SD, n = 3). **P < 0.01 vs Control; □□P < 0.01 vs Free PTX; ▵▵P < 0.01 vs 7pep-M-PTX. (B) Effects of different treatments with nanomedicines on the ratios of LC3-II to LC3-I. Levels of LC3B proteins expression were measured by western-blot, and the ratios of LC3-II to LC3-I were calculated by comparing the band densities (mean ± SD, n = 3). *P < 0.05 vs Control. (C) Images and quantitative analysis of colocalization of the LC3B-labeled autophagic vesicles (green dots) and Lyso-tracker red-labeled lysosomes (red). The white scales represent 10 μm. Yellow-framed inserts show the scatter plots generated by Image J software (mean ± SD, n = 6). *P < 0.05 vs Control; ▵▵P < 0.01 vs 7pep-M-PTX. (D) Images and quantitative analysis of autophagic vacuoles specifically labeled by Cyto-ID dye (green). The cell nuclei were stained with Hoechst 33342 (blue). The white scales represent 10 μm. Each bar on the histogram represents mean fluorescence intensity obtained from 6 randomly selected cells (mean ± SD, n = 6).