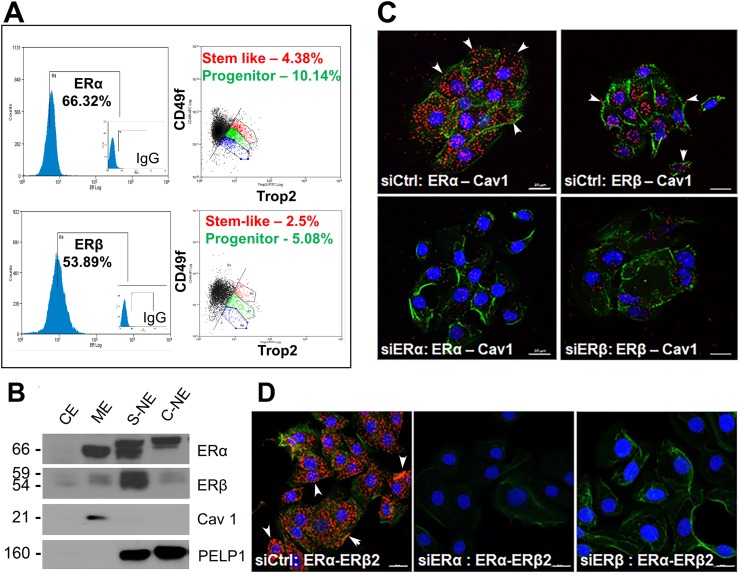
Figure 1.
Localization of ERα and ERβ at the membrane of three-dimensional prostaspheres (PSs). (A) PSs cultured for 7 d were dispersed, triple stained for stem markers CD49f, Trop2, and either ERα or ERβ, and sorted by flow cytometry. Triple-stained subpopulations of cells comprising the stem cell and early-stage progenitor subpopulation expressing either ERα or ERβ were identified using the Summit software analysis package (n = 4). (B) Subcellular protein fractionation of day 7 PSs revealed the presence of ERα and ERβ in the membrane extract (ME), the soluble nuclear extract (S-NE), and chromatin-bound nuclear extract (C-NE), with limited protein found in the soluble cytoplasmic extract (CE). Caveolin-1 (Cav1) was used as a membrane marker, and PELP1 was used as a nuclear marker (n = 3). Differential ER migration between fractions is likely due to use of different extraction buffers and/or posttranslational modifications. (C) PLA on day 5 PSs revealed ERα and ERβ interactions with Cav1 at the cell membrane (arrowheads point to examples) and in cytoplasm, presumably during transport or turnover. Loss of most signal with ERα or ERβ knockdown by siRNA (bottom panels) documents signal specificity. (D) PLA on day 5 PSs between ERα and ERβ, depicted by the red punctate staining, indicates ERα–ERβ heterodimerization, including at the membrane (arrowheads). Loss of signals with siRNA for either ER documents specificity. For both (C) and (D), Alexa Fluor 488–phalloidin stain was used to visualize the membrane and demarcate cells. Nuclei are demarcated by DAPI counterstain (blue). Scale bars, 20 µm. Each PLA was repeated for n = 3.