ABSTRACT
In Arabidopsis, the floral meristem is essential for the production of floral organs. The floral meristem is initially maintained to contribute cells for floral organ formation. However, this stem cell activity needs be completely terminated at a certain floral developmental stage to ensure the proper development of floral reproductive organs. Here, we have reviewed recent findings on the complex regulation of floral meristem activities, which involve signaling cascades, transcriptional regulation, epigenetic mechanisms and hormonal control for floral meristem determinacy in Arabidopsis.
KEYWORDS: arabidopsis, floral meristem, WUS, determinacy, flower development
Introduction
In Arabidopsis, a fixed number of floral organs are generated by floral meristems. Floral meristems are established by the homeobox gene WUSCHEL (WUS) expressed in the cells of the organizing center (OC). The WUS protein specifies stem cell identity in a non-cell autonomous manner by migrating from the OC into the central zone (CZ).1 In the CZ, WUS binds directly to the CLAVATA3 (CLV3) locus and activates CLV3, which is uniquely expressed in aerial stem cells.1 The CLV3 peptide diffuses to the OC region, can be recognized by the CLAVATA (CLV) receptor system, including CLV1, CLV2, CORYNE (CRN), BARELY ANY MERISTEMS (BAMs) and CLAVATA3 INSENSITIVE RECEPTOR KINASE (CIKs), and restricts WUS expression.2–6 In early stages of flower development, the CLV-WUS signaling pathway maintains stem cell homeostasis, thus giving rise to proper flower formation.7 However, stem cell activity is terminated at floral stage 68 to ensure initiation and proper development of carpels.
In this review, we mainly summarize the mechanisms of floral stem cell termination. These mechanisms involve transcriptional and epigenetic regulation of multiple factors. Phytohormones also play an indispensable role in the intricate and precise control of floral meristem determinacy.
AGAMOUS (AG) and SUPERMAN (SUP) repress WUS from early floral stages
AG directly represses WUS from floral stage 3
The floral homeotic gene AG alone specifies carpel identity in Arabidopsis.9 In addition, AG is a critical transcription factor involved in the direct repression of WUS in the floral meristem. AG activity is induced at the center of stage 3 flower buds by WUS and LEAFY (LFY).10,11 In turn, AG represses WUS expression by directly binding to the locus from floral stage 3 onward.12 Polycomb group protein (PcG) complex, which can introduce the repressive mark, histone H3 Lys 27 trimethylation (H3K27me3) to silence genes,13 is also involved in WUS regulation by AG. Polycomb repressive complex 1 (PRC1) factor TERMINAL FLOWER 2 (TFL2) is recruited by AG to the WUS locus for direct repression. The PRC2 factor, CURLY LEAF (CLF), is also required for the regulation of AG-mediated FM determinacy. A recent study has shown that there is a chromatin loop on WUS that is formed by the interaction between AG and TFL2 at two specific regions on the WUS locus.14 The recruitment of RNA polymerase II is blocked by this chromatin loop on WUS, and WUS expression is thereby repressed by the loop. However, WUS is only mildly repressed by AG from floral stage 3 onward. Although the null ag-1 mutant shows loss of floral meristem determinacy with the homeotic transformation of flower phenotype to sepal-petal-petal reiteration,15 transgenic lines of 35S:AG, in which AG is over-expressed, still produce flowers with normal carpels.16 This finding suggests that floral meristem termination requires factors other than the AG protein alone.
SUPERMAN represses WUS from early stages of flower development
SUPERMAN (SUP) encodes a C2H2-type zinc finger transcription factor, and the loss of SUP function leads to floral meristem indeterminacy and over-production of stamens at the expense of proper carpel formation.17 A recent paper found that extra stamens in sup mutants originate from cells in both third and fourth whorls and undergo fate changes from carpels to stamens.18 In loss-of-function sup, ag and clv3 triple mutant flowers, the floral meristem becomes much larger than the meristem in ag clv3 double mutants, suggesting SUP may regulate floral stem cell activity in parallel with AG and CLV3.19 At floral stage 3, SUP expression is observed on both sides of the boundary between whorl 3 stamens and whorl 4 carpels.17,18 SUP forms a repressor complex with PcG factors CURLY LEAF (CLF) and TFL2 and fine-tunes local auxin signaling by negatively regulating the expression of the auxin synthesis genes YUCCA1/4 (YUC1/4).20 In sup mutant flowers, auxin production is increased in the whorl 3/4 boundary region, while auxin accumulation is reduced in the center of the flowers.20 Repression of auxin biosynthesis by SUP is pivotal for floral meristem determinacy from floral stage 3 onward; at this stage the stem cell marker CLV3-GFP begins to indicate there is an increase in stem cell numbers in sup mutant flowers compared to wild type flowers.20,21
AG induces KNUCKLES (KNU) and CRABS CLAW (CRC) to repress WUS at floral stage 6
AG induces KNU to terminate floral stem cells
WUS expression is directly repressed by AG from floral stage 3 onward.12 However, this inhibition is moderate and insufficient to terminate stem cell activity. At floral stage 6, AG directly induces KNUCKLES (KNU), which encodes a C2H2-type zinc finger protein. Both weak mutant knu-1 and null mutant knu-2 display indeterminate floral phenotypes with ectopic reiterative stamens and carpels formed within gynecium due to prolonged WUS activity.22,23 In contrast, transgenic plants with an over-expression of KNU produce flowers resembling the loss-of-function mutant wus-1 phenotype; this observation suggests that KNU plays a decisive role for WUS termination at stage 6.8,23,24
From floral stage 3, AG directly binds to the KNU promoter.23 However, KNU expression is not activated immediately after AG binding. There is a characteristic 153 bp Polycomb Responsive Element (PRE) on the KNU promoter; PRE formation coincides with AG binding to CArG boxes.24 The PRC2 factors, including EMBRYONIC FLOWER 2 (EMF2) and FERTILIZATION INDEPENDENT ENDOSPERM (FIE), associated with KNU PRE, are displaced by AG from floral stage 3; this action leads to the failure of the maintenance of repressive marker H3K27me3 on KNU chromatin.24 Through 1 ~ 2 rounds of cell division, which takes approximately 2 days,24 H3K27me3 repression on KNU is diluted. Therefore, KNU expression is activated at floral stage 6. The timing of KNU induction is pivotal for normal floral development. Delayed or early KNU expression leads to indeterminate floral organs or to flowers without carpels, respectively.7
The termination of floral stem cells is characterized by the silencing of WUS activity at floral stage 6, which is a multistep process mediated by the function of KNU.8 As a repressor, KNU directly binds to the WUS proximal promoter region and co-localizes to the SPLAYED (SYD) binding site. SYD is a SWI/SNF chromatin remodeling factor and functions as a key activator of WUS.25 KNU binding causes SYD eviction, reduces DNA accessibility of the WUS locus, and decreases the levels of active H3K4me3 histone markers and H3 acetylation on WUS chromatin. These events are associated with the repression of WUS mRNA within 4 hours of KNU activation.8
The deacetylation of histones is reported to be required for WUS repression.8,26 As an initial step, the adaptor protein MINI ZINC FINGER2 (MIF2) binds to the first WUS intron and recruits KNU, TOPLESS (TPL) and HISTONE DEACETYLASE19 (HDA19) to form a transcriptional repressor complex that represses WUS.26 This activity possibly occurs simultaneous with the eviction of SYD.
Next, KNU interacts with FIE and recruits PRC2 to WUS in a KNU-dependent manner; this recruitment leads to increased H3K27me3 levels on WUS chromatin approximately 8 hours after KNU activity is induced.8 The deposition of H3K27me3 is only detected several hours later than WUS transcriptional repression and reduction of active markers on WUS chromatin. The deposition of H3K27me3 may be a prerequisite step for PcG-mediated silencing of WUS. In the transgenic co-suppression line 35S:GFP-FIE, which has mostly silenced FIE activity,27 ectopic carpelloid tissue is found inside the gynecium; this is the result of prolonged WUS activity.8 On the other hand, over-expression of KNU in tfl2 and clf mutants still gives rise to normal carpels, unlike KNU over-expression in wild types in which carpel formation is fully abolished. All these findings suggest that PcG activity is required to silence WUS. Hence, KNU integrates transcriptional repression and epigenetic silencing of WUS through multiple steps for floral stem cell termination.
AG induces CRC for floral meristem termination through fine-tuning auxin homeostasis
CRC encodes a YABBY family transcription factor, which is directly activated by AG at floral stage 5 ~ 6.28–30 Single mutants of crc have no obvious floral meristem defects; however, stronger floral meristem indeterminacy is observed in crc-1 knu-1 double mutants compared to knu-1 mutants, indicating that CRC is also involved in the regulation of floral meristem determinacy.31,32 At floral stage 6, CRC and AG synergistically activate YUC4 leading to auxin accumulation in the floral meristem.33 CRC can also directly repress TORNADO2 (TRN2), which encodes a transmembrane protein involved in the negative regulation of auxin signaling.34–37 To establish the auxin maxima needed to direct floral organ differentiation in the floral meristem, CRC finely modulates auxin homeostasis by both promoting YUC4 and repressing TRN2.30
AG activity is modulated by SEPALLATA3 (SEP3) and APETALA2 (AP2)
AG plays dual roles in floral organ identity control, and floral meristem regulation, but it is still unknown whether AG participates in different complexes due to its distinct functions. It has recently been reported that the MADS-domain protein complex formed by AG and SEPALLATA3 (SPE3) tetramers is required to activate AG’s direct downstream genes KNU and CRC for floral meristem determinacy.38
Interestingly, the role of AG in floral stem cell regulation is antagonized by the floral homeotic protein APETALA2 (AP2), which represses KNU expression at floral stage 6.39 Therefore, AP2 could serve as a brake in the feed-forward regulatory loop consisting of AG, KNU and WUS.
Other factors involved in floral meristem regulation
In flower development, auxin and cytokinin interact to co-regulate floral meristem determinacy. Promoted by auxin, AUXIN RESPONSE FACTOR3 (ARF3) functions in two ways to inhibit floral stem cell activity. ARF3 can directly bind to the WUS promoter to repress its expression.40 ARF3 can also repress cytokinin activity by repressing the ISOPENTENYLTRANSFERASE (IPT) and LONELY GUY (LOG) family genes that encode enzymes for cytokinin biosynthesis.41 Furthermore, ARF3 directly represses the expression of ARABIDOPSIS HISTIDINE KINASE4 (AHK4), which encodes a cytokinin receptor; this repression leads to a decrease in cytokinin activity and WUS repression.41 In flower development, both AG and AP2 can dynamically regulate ARF3 expression,40,42 thereby linking ARF3 transcription factor activities with phytohormones for the regulation of floral stem cells.
At floral stage 3, AG expression is induced at the center of flower buds by LFY and WUS.10,11 ULTRAPETALA1 (ULT1), a trxG protein, regulates floral stem cell activity by inducing AG expression in a LFY-independent manner.43 PERIANTHIA (PAN) is a bZIP transcription factor, and the expression region of PAN overlaps with AG; PAN can also directly activate AG expression.44,45 A reduced AG expression level and an increased number of floral organs are observed in pan mutant flowers.45 In addition, other genes, including SQUINT (SQN) and REBELOTE (RBL), function redundantly upstream of AG and maintain AG expression from floral stage 3 onward.32
In shoot apical meristems and floral meristems, a WUS-independent pathway can also control stem cell activity.46 The stem cell activity is repressed by HD-zip III transcription factors, including PHABULOSA (PHB), PHAVOLUTA (PHV) and CORONA (CNA). Premature termination of the floral meristem is partially rescued in wus phb phv cna quadruple mutants. PHB, PHV, CNA are the targets of miR165/166 that has been repressed by ARGONAUTE10 (AGO10).47 Another factor, REVOLUTA (REV) is required for floral meristem specification; REV antagonizes the function of PHB/PHV/CAN.48,49 In addition, WUS expression is repressed by an ERECTA (ER)- and JABBA (JAB)-mediated signaling pathway, independent of the CLV pathway.50 It is reported that WUS expression levels significantly increase in jba-1D/+ er-20 compared to jba-1D/+.50
In most events, the key for floral meristem determinacy is the repression and silencing of WUS, a central player in the establishment and maintenance of stem cell activity. Meanwhile, the repression of another key gene, CLV3, may also be necessary for floral meristem determinacy. It was reported that CLV3 expression is transcriptionally repressed by FAR-RED ELONGATED HYPOCOTYL3 (FHY3) in flower development.51 Notably, repression of CLV3 is observed within 4 hours and is independent of cycloheximide activity when KNU is expressed; this result indicates that KNU may directly repress both CLV3 and WUS at floral stage 6, thus ensuring the termination of robust floral stem cell activities within a narrow window of time.8
Conclusions and future perspectives
Floral meristem determinacy is governed by a complex regulatory network (Figure 1). In this network, AG-mediated downstream regulatory pathways play a central role. The activation of KNU to silence WUS plays a pivotal role for floral meristem termination. This AG-KNU-WUS pathway also involves epigenetic events including the dynamic eviction of PcG from KNU and the recruitment of PcG to WUS. In addition, plant hormones regulated by SUP and CRC play important roles in balancing floral stem cell proliferation and differentiation. Because of the complex nature of floral stem cell regulation, many other factors are yet to be discovered. Whether these regulatory mechanisms are conserved in other plant species are intriguing questions whose answers will shed light on the enhancement of future crop yields.
Figure 1.
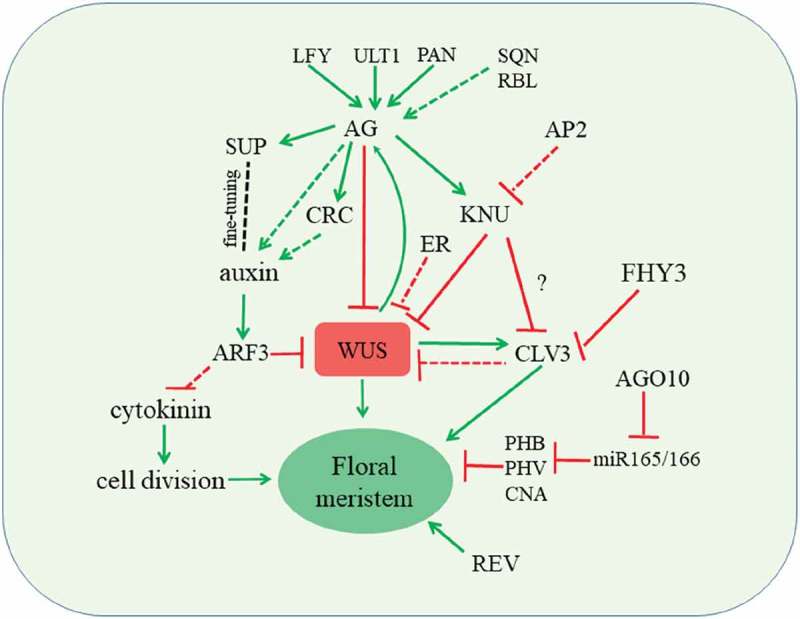
Termination of floral stem cell activity is controlled by multiple pathways. Red and green lines indicate repressions and activations, respectively. Solid and dashed lines indicate direct and indirect regulation, respectively.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31670308].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- 1.Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV.. WUSCHEL protein movement mediates stem cell homeostasis in the arabidopsis shoot apex. Genes Dev. 2011;25:1–5. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R.. Dependence of stem cell fate in arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 3.Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 4.Hazak O, Hardtke CS. CLAVATA 1-type receptors in plant development. J Exp Bot. 2016;67:4827–4833. doi: 10.1093/jxb/erw247. [DOI] [PubMed] [Google Scholar]
- 5.Janocha D, Lohmann JU. From signals to stem cells and back again. Curr Opin Plant Biol. 2018;45:136–142. doi: 10.1016/j.pbi.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu C, Zhu Y, Cui Y, Cheng K, Liang W, Wei Z, Zhu M, Yin H, Zeng L, Xiao Y, et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat Plants. 2018;4:205. doi: 10.1038/s41477-018-0123-z. [DOI] [PubMed] [Google Scholar]
- 7.Sun B, Ito T. Regulation of floral stem cell termination in arabidopsis. Front Plant Sci. 2015;6:17. doi: 10.3389/fpls.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun B, Zhou Y, Cai J, Shang E, Yamaguchi N, Xiao J, Looi L-S, Wee W-Y, Gao X, Wagner D, et al. Integration of transcriptional repression and Polycomb-mediated silencing of WUSCHEL in floral meristems. Plant Cell. tpc. 00450.02018 2019. doi: 10.1105/tpc.18.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- 10.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 11.Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Kim YJ, Müller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X. AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell. 2011;23:3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafos M, Carty CL, Taylor K, Schumacher FR, Hindorff LA, Ambite JL, Anderson G, Best LG, Brown-Gentry K, Bůžková P, et al. Dynamic regulation of H3K27 trimethylation during arabidopsis differentiation. PLoS Genet. 2011;7:e1002040. doi: 10.1371/journal.pgen.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo L, Cao X, Liu Y, Li J, Li Y, Li D, Zhang K, Gao C, Dong A, Liu X. A chromatin loop represses WUSCHEL expression in arabidopsis. Plant J. 2018;94:1083–1097. doi: 10.1111/tpj.13921. [DOI] [PubMed] [Google Scholar]
- 15.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizukami Y, Ma H. Determination of arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. SUPERMAN, a regulator of floral homeotic genes in arabidopsis. Development. 1992;114:599–615. [DOI] [PubMed] [Google Scholar]
- 18.Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of arabidopsis thaliana flowers. Proc Natl Acad Sci. 2017;114:7166–7171. doi: 10.1073/pnas.1705977114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uemura A, Yamaguchi N, Xu Y, Wee W, Ichihashi Y, Suzuki T, Shibata A, Shirasu K, Ito T. Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in arabidopsis. Sex Plant Reprod. 2018;31:89–105. doi: 10.1007/s00497-017-0315-0. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Prunet N, Gan E-S, Wang Y, Stewart D, Wellmer F, Huang J, Yamaguchi N, Tatsumi Y, Kojima M, et al. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. Embo J. 2018;37. doi: 10.15252/embj.201797499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Yamaguchi N, Gan E, Ito T. When to stop: an update on molecular mechanisms of floral meristem termination. J Exp Bot. 2019;70:1711–1718. doi: 10.1093/jxb/erz048. [DOI] [PubMed] [Google Scholar]
- 22.Payne T, Johnson SD, Koltunow AMG. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the arabidopsis gynoecium. Development. 2004;131:3737–3749. doi: 10.1242/dev.01216. [DOI] [PubMed] [Google Scholar]
- 23.Sun B, Xu Y, Ng K, Ito T. A timing mechanism for stem cell maintenance and differentiation in the arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun B, Looi L-S, Guo S, He Z, Gan E-S, Huang J, Xu Y, Wee W-Y, Ito T. Timing mechanism dependent on cell division is invoked by polycomb eviction in plant stem cells. Science. 2014;343:1248559. doi: 10.1126/science.1248559. [DOI] [PubMed] [Google Scholar]
- 25.Kwon CS, Chen C, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollier N, Sicard A, Leblond J, Latrasse D, Gonzalez N, Gévaudant F, Benhamed M, Raynaud C, Lenhard M, Chevalier C, et al. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in arabidopsis and tomato. Plant Cell. 2018;30:83–100. doi: 10.1105/tpc.17.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–719. [DOI] [PubMed] [Google Scholar]
- 28.Gomezmena C, De Folter S, Costa MMR, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Baum SF, Alvarez J, Patel A, Chitwood DH, Bowman JL. Activation of CRABS CLAW in the nectaries and carpels of arabidopsis. Plant Cell. 2005;17:25–36. doi: 10.1105/tpc.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi N, Huang J, Xu Y, Tanoi K, Ito T. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat Commun. 2017;8:1125. doi: 10.1038/s41467-017-01252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breuil-Broyer S, Trehin C, Morel P, Boltz V, Sun B, Chambrier P, Ito T, Negrutiu I. Analysis of the arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male–female boundary, flower meristem termination and carpel compartmentalization. Ann Bot. 2016;117:905–923. doi: 10.1093/aob/mcw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prunet N, Morel P, Thierry A-M, Eshed Y, Bowman JL, Negrutiu I, Trehin C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in arabidopsis thaliana. Plant Cell. 2008;20:901–919. doi: 10.1105/tpc.107.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi N, Huang J, Tatsumi Y, Abe M, Sugano SS, Kojima M, Takebayashi Y, Kiba T, Yokoyama R, Nishitani K, et al. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat Commun. 2018;9:5290. doi: 10.1038/s41467-018-07763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boavida LC, Qin P, Broz M, Becker JD, McCormick S. Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 2013;163:696–712. doi: 10.1104/pp.113.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Muto A, Van de Velde J, Neyt P, Himanen K, Vandepoele K, Van Lijsebettens M. Functional analysis of the arabidopsis TETRASPANIN gene family in plant growth and development. Plant Physiol. 2015;169:2200–2214. doi: 10.1104/pp.15.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu W, Chandler JW, Cnops G, Van Lijsebettens M, Werr W. Mutations in the TORNADO2 gene affect cellular decisions in the peripheral zone of the shoot apical meristem of arabidopsis thaliana. Plant Mol Biol. 2007;63:731–744. doi: 10.1007/s11103-006-9105-z. [DOI] [PubMed] [Google Scholar]
- 37.Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, Malenica N, Luschnig C, Tietz O, Ditengou F, Palme K, et al. The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in arabidopsis thaliana. Plant Cell. 2006;18:852–866. doi: 10.1105/tpc.105.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hugouvieux V, Silva CS, Jourdain A, Stigliani A, Charras Q, Conn V, Conn SJ, Carles CC, Parcy F, Zubieta C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in arabidopsis. Nucleic Acids Res. 2018;46:4966–4977. doi: 10.1093/nar/gky205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z, Shi T, Zheng B, Yumul RE, Liu X, You C, Gao Z, Xiao L, Chen X. APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in arabidopsis thaliana. New Phytol. 2017;215:1197–1209. doi: 10.1111/nph.14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Dinh TT, Li D, Shi B, Li Y, Cao X, Guo L, Pan Y, Jiao Y, Chen X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014;80:629–641. doi: 10.1111/tpj.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K, Wang R, Zi H, Li Y, Cao X, Li D, Guo L, Tong J, Pan Y, Jiao Y, et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell. 2018;30:324–346. doi: 10.1105/tpc.17.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng K, Yu H, Ito T, Weigel D. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 2009;7:e1000251. doi: 10.1371/journal.pbio.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carles CC, Fletcher JC. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009;23:2723–2728. doi: 10.1101/gad.1812609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das P, Ito T, Wellmer F, Vernoux T, Dedieu A, Traas J, Meyerowitz EM. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development. 2009;136:1605–1611. doi: 10.1242/dev.035436. [DOI] [PubMed] [Google Scholar]
- 45.Maier AT, Stehling-Sun S, Wollmann H, Demar M, Hong RL, Haubeiss S, Weigel D, Lohmann JU. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development. 2009;136:1613–1620. doi: 10.1242/dev.033647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C, Clark SE, Candela H. A WUSCHEL-independent stem cell specification pathway is repressed by PHB, PHV and CNA in arabidopsis. PLoS One. 2015;10:e0126006. doi: 10.1371/journal.pone.0126006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 48.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. [DOI] [PubMed] [Google Scholar]
- 50.Mandel T, Moreau F, Kutsher Y, Fletcher JC, Carles CC, Eshed Williams L. The ERECTA receptor kinase regulates arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development. 2014;141:830–841. doi: 10.1242/dev.104687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D, Fu X, Guo L, Huang Z, Li Y, Liu Y, He Z, Cao X, Ma X, Zhao M, et al. FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in arabidopsis. Proc Natl Acad Sci U S A. 2016;113:9375–9380. doi: 10.1073/pnas.1602960113. [DOI] [PMC free article] [PubMed] [Google Scholar]


